Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing
- PMID: 25144201
- PMCID: PMC4140814
- DOI: 10.1371/journal.pone.0105592
Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing
Abstract
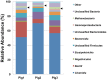
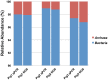
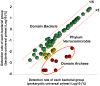
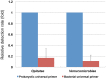
For the analysis of microbial community structure based on 16S rDNA sequence diversity, sensitive and robust PCR amplification of 16S rDNA is a critical step. To obtain accurate microbial composition data, PCR amplification must be free of bias; however, amplifying all 16S rDNA species with equal efficiency from a sample containing a large variety of microorganisms remains challenging. Here, we designed a universal primer based on the V3-V4 hypervariable region of prokaryotic 16S rDNA for the simultaneous detection of Bacteria and Archaea in fecal samples from crossbred pigs (Landrace × Large white × Duroc) using an Illumina MiSeq next-generation sequencer. In-silico analysis showed that the newly designed universal prokaryotic primers matched approximately 98.0% of Bacteria and 94.6% of Archaea rRNA gene sequences in the Ribosomal Database Project database. For each sequencing reaction performed with the prokaryotic universal primer, an average of 69,330 (± 20,482) reads were obtained, of which archaeal rRNA genes comprised approximately 1.2% to 3.2% of all prokaryotic reads. In addition, the detection frequency of Bacteria belonging to the phylum Verrucomicrobia, including members of the classes Verrucomicrobiae and Opitutae, was higher in the NGS analysis using the prokaryotic universal primer than that performed with the bacterial universal primer. Importantly, this new prokaryotic universal primer set had markedly lower bias than that of most previously designed universal primers. Our findings demonstrate that the prokaryotic universal primer set designed in the present study will permit the simultaneous detection of Bacteria and Archaea, and will therefore allow for a more comprehensive understanding of microbial community structures in environmental samples.
Conflict of interest statement
Figures
References
-
- Zumsteg A, Luster J, Goransson H, Smittenberg RH, Brunner I, et al. (2012) Bacterial, archaeal and fungal succession in the forefield of a receding glacier. Microb Ecol 63: 552–564. - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

