Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity
- PMID: 25144274
- PMCID: PMC4140854
- DOI: 10.1371/journal.ppat.1004321
Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity
Abstract
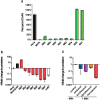
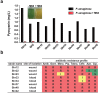
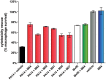
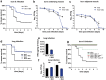
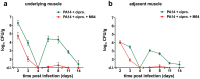
Etiological agents of acute, persistent, or relapsing clinical infections are often refractory to antibiotics due to multidrug resistance and/or antibiotic tolerance. Pseudomonas aeruginosa is an opportunistic Gram-negative bacterial pathogen that causes recalcitrant and severe acute chronic and persistent human infections. Here, we target the MvfR-regulated P. aeruginosa quorum sensing (QS) virulence pathway to isolate robust molecules that specifically inhibit infection without affecting bacterial growth or viability to mitigate selective resistance. Using a whole-cell high-throughput screen (HTS) and structure-activity relationship (SAR) analysis, we identify compounds that block the synthesis of both pro-persistence and pro-acute MvfR-dependent signaling molecules. These compounds, which share a benzamide-benzimidazole backbone and are unrelated to previous MvfR-regulon inhibitors, bind the global virulence QS transcriptional regulator, MvfR (PqsR); inhibit the MvfR regulon in multi-drug resistant isolates; are active against P. aeruginosa acute and persistent murine infections; and do not perturb bacterial growth. In addition, they are the first compounds identified to reduce the formation of antibiotic-tolerant persister cells. As such, these molecules provide for the development of next-generation clinical therapeutics to more effectively treat refractory and deleterious bacterial-human infections.
Conflict of interest statement
LR is the scientific founder, consultant and scientific advisory board member of Spero Therapeutics LLC. No funding from Spero Therapeutics was received. This does not alter our adherence to all PLOS policies on sharing data and materials.
Figures




References
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, et al. (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48: 1–12. - PubMed
-
- Livermore DM (2012) Fourteen years in resistance. Int J Antimicrob Agents 39: 283–294. - PubMed
-
- Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, et al. (2008) The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46: 155–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

