Specificity and affinity quantification of flexible recognition from underlying energy landscape topography
- PMID: 25144525
- PMCID: PMC4140643
- DOI: 10.1371/journal.pcbi.1003782
Specificity and affinity quantification of flexible recognition from underlying energy landscape topography
Abstract
Flexibility in biomolecular recognition is essential and critical for many cellular activities. Flexible recognition often leads to moderate affinity but high specificity, in contradiction with the conventional wisdom that high affinity and high specificity are coupled. Furthermore, quantitative understanding of the role of flexibility in biomolecular recognition is still challenging. Here, we meet the challenge by quantifying the intrinsic biomolecular recognition energy landscapes with and without flexibility through the underlying density of states. We quantified the thermodynamic intrinsic specificity by the topography of the intrinsic binding energy landscape and the kinetic specificity by association rate. We found that the thermodynamic and kinetic specificity are strongly correlated. Furthermore, we found that flexibility decreases binding affinity on one hand, but increases binding specificity on the other hand, and the decreasing or increasing proportion of affinity and specificity are strongly correlated with the degree of flexibility. This shows more (less) flexibility leads to weaker (stronger) coupling between affinity and specificity. Our work provides a theoretical foundation and quantitative explanation of the previous qualitative studies on the relationship among flexibility, affinity and specificity. In addition, we found that the folding energy landscapes are more funneled with binding, indicating that binding helps folding during the recognition. Finally, we demonstrated that the whole binding-folding energy landscapes can be integrated by the rigid binding and isolated folding energy landscapes under weak flexibility. Our results provide a novel way to quantify the affinity and specificity in flexible biomolecular recognition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
 is the fraction of native interfacial binding contacts. "Ind" and "Eff" are the abbreviations for "Independent" and "Effective" binding, respectively.
is the fraction of native interfacial binding contacts. "Ind" and "Eff" are the abbreviations for "Independent" and "Effective" binding, respectively.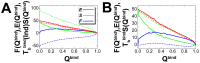
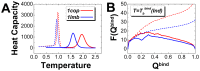
 ), and (B) corresponding rigid and flexible binding transition temperature (
), and (B) corresponding rigid and flexible binding transition temperature ( ). Energy, entropy and free energy are in reduced unit.
). Energy, entropy and free energy are in reduced unit.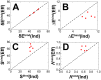
 , (B) energy roughness
, (B) energy roughness  , (C) entropy
, (C) entropy  and (D) energy landscape topography measure
and (D) energy landscape topography measure  .
.
 . (B) The differences of kinetics, represented by the ratio of binding time between rigid and flexible binding, are plotted along the differences of intrinsic specificity between rigid and flexible binding.
. (B) The differences of kinetics, represented by the ratio of binding time between rigid and flexible binding, are plotted along the differences of intrinsic specificity between rigid and flexible binding.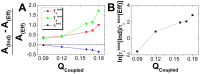
 , where
, where  and
and  are the quantities of the rigid and flexible binding. The quantities
are the quantities of the rigid and flexible binding. The quantities  are
are  ,
,  and
and  , corresponding to the binding transition temperature, glassy trapping temperature and binding energy landscape topography measure, respectively. (B) The differences of kinetics, represented by the ratio of association time between rigid and flexible binding, are plotted as a function of
, corresponding to the binding transition temperature, glassy trapping temperature and binding energy landscape topography measure, respectively. (B) The differences of kinetics, represented by the ratio of association time between rigid and flexible binding, are plotted as a function of  .
.  describes the degree of interfacial flexibility.
describes the degree of interfacial flexibility.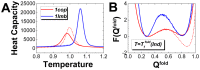
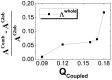
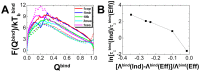
 , where
, where  and
and  are the energy landscape topography measures
are the energy landscape topography measures  of the combined and whole global binding-folding. "Comb" and "Glob" are the abbreviations for "combined" and "whole global" binding-folding, respectively.
of the combined and whole global binding-folding. "Comb" and "Glob" are the abbreviations for "combined" and "whole global" binding-folding, respectively.References
-
- Otlewski J, Apostoluk W (1997) Structural and energetic aspects of protein-protein recognition. Acta Biochimica Polonica 44: 367–387. - PubMed
-
- Muller-Dethlefs K, Hobza P (2000) Noncovalent interactions: A challenge for experiment and theory. Chem Rev 100: 143–167. - PubMed
-
- Fischer E (1894) Einfluss der configuration auf die wirkung der enzyme. Ber Dtsch Chem Ges 27: 2984–2993.
-
- McCammon JA (1998) Theory of biomolecular recognition. Curr Opin Struct Biol 8: 245–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

