Commensal-pathogen interactions in the intestinal tract: lactobacilli promote infection with, and are promoted by, helminth parasites
- PMID: 25144609
- PMCID: PMC4822684
- DOI: 10.4161/gmic.32155
Commensal-pathogen interactions in the intestinal tract: lactobacilli promote infection with, and are promoted by, helminth parasites
Abstract
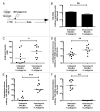
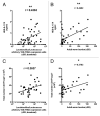
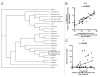
The intestinal microbiota are pivotal in determining the developmental, metabolic and immunological status of the mammalian host. However, the intestinal tract may also accommodate pathogenic organisms, including helminth parasites which are highly prevalent in most tropical countries. Both microbes and helminths must evade or manipulate the host immune system to reside in the intestinal environment, yet whether they influence each other's persistence in the host remains unknown. We now show that abundance of Lactobacillus bacteria correlates positively with infection with the mouse intestinal nematode parasite, Heligmosomoides polygyrus, as well as with heightened regulatory T cell (Treg) and Th17 responses. Moreover, H. polygyrus raises Lactobacillus species abundance in the duodenum of C57BL/6 mice, which are highly susceptible to H. polygyrus infection, but not in BALB/c mice, which are relatively resistant. Sequencing of samples at the bacterial gyrB locus identified the principal Lactobacillus species as L. taiwanensis, a previously characterized rodent commensal. Experimental administration of L. taiwanensis to BALB/c mice elevates regulatory T cell frequencies and results in greater helminth establishment, demonstrating a causal relationship in which commensal bacteria promote infection with an intestinal parasite and implicating a bacterially-induced expansion of Tregs as a mechanism of greater helminth susceptibility. The discovery of this tripartite interaction between host, bacteria and parasite has important implications for both antibiotic and anthelmintic use in endemic human populations.
Keywords: Commensal; Duodenum; Lactobacillus; Microbiota; Nematode; Th17; Treg.
Figures


References
-
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. PMID:19343057; http://dx.doi.org/10.1038/nri2515. - DOI - PMC - PubMed
-
- Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol. 2012;24:58–66. PMID:22178452; http://dx.doi.org/10.1016/j.smim.2011.11.008. - DOI - PMC - PubMed
-
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. PMID:22674334; http://dx.doi.org/10.1126/science.1223490. - DOI - PMC - PubMed
-
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–84. PMID:23778795; http://dx.doi.org/10.1038/ni.2640. - DOI - PMC - PubMed
-
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. PMID:22699609; http://dx.doi.org/10.1038/nature11234. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
