An evolutionary preserved intergenic spacer in gadiform mitogenomes generates a long noncoding RNA
- PMID: 25145347
- PMCID: PMC4236577
- DOI: 10.1186/s12862-014-0182-3
An evolutionary preserved intergenic spacer in gadiform mitogenomes generates a long noncoding RNA
Abstract
Background: Vertebrate mitogenomes are economically organized and usually lack intergenic sequences other than the control region. Intergenic spacers located between the tRNA(Thr) and tRNA(Pro) genes ("T-P spacers") have been observed in several taxa, including gadiform species, but information about their biological roles and putative functions is still lacking.
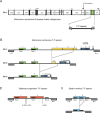
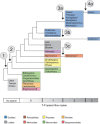
Results: Sequence characterization of the complete European hake Merluccius merluccius mitogenome identified a complex T-P spacer ranging in size from 223-532 bp. Further analyses of 32 gadiform species, representing 8 families and 28 genera, revealed the evolutionary preserved presence of T-P spacers across all taxa. Molecular complexity of the T-P spacers was found to be coherent with the phylogenetic relationships, supporting a common ancestral origin and gain of function during codfish evolution. Intraspecific variation of T-P spacer sequences was assessed in 225 Atlantic cod specimens and revealed 26 haplotypes. Pyrosequencing data representing the mito-transcriptome poly (A) fraction in Atlantic cod identified an abundant H-strand specific long noncoding RNA of about 375 nt. The T-P spacer corresponded to the 5' part of this transcript, which terminated within the control region in a tail-to-tail configuration with the L-strand specific transcript (the 7S RNA).
Conclusions: The T-P spacer is inferred to be evolutionary preserved in gadiform mitogenomes due to gain of function through a long noncoding RNA. We suggest that the T-P spacer adds stability to the H-strand specific long noncoding RNA by forming stable hairpin structures and additional protein binding sites.
Figures


References
-
- Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. - PubMed
-
- Brown C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu Rev Ecol Syst. 1987;18:269–292.
-
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol Evol. 2006;23:227–234. - PubMed
-
- Kumazawa Y, Ota H, Nishida M, Ozawa T. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 1996;13:1242–1254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

