RNAi screening comes of age: improved techniques and complementary approaches
- PMID: 25145850
- PMCID: PMC4204798
- DOI: 10.1038/nrm3860
RNAi screening comes of age: improved techniques and complementary approaches
Abstract
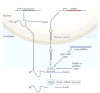
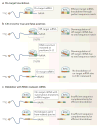
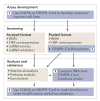
Gene silencing through sequence-specific targeting of mRNAs by RNAi has enabled genome-wide functional screens in cultured cells and in vivo in model organisms. These screens have resulted in the identification of new cellular pathways and potential drug targets. Considerable progress has been made to improve the quality of RNAi screen data through the development of new experimental and bioinformatics approaches. The recent availability of genome-editing strategies, such as the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 system, when combined with RNAi, could lead to further improvements in screen data quality and follow-up experiments, thus promoting our understanding of gene function and gene regulatory networks.
Conflict of interest statement
Figures
References
-
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Rev Mol Cell Biol. 2007;8:23–36. - PubMed
-
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nature Rev Genetics. 2008;9:554–566. - PubMed
-
- Grimm S. The art and design of genetic screens: mammalian culture cells. Nature reviews Genetics. 2004;5:179–189. - PubMed
-
- Zhuang JJ, Hunter CP. RNA interference in Caenorhabditis elegans: uptake, mechanism, and regulation. Parasitology. 2012;139:560–573. - PubMed
-
- Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nature Methods. 2006;3:701–706. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

