Host lifestyle affects human microbiota on daily timescales
- PMID: 25146375
- PMCID: PMC4405912
- DOI: 10.1186/gb-2014-15-7-r89
Host lifestyle affects human microbiota on daily timescales
Erratum in
-
Erratum to: Host lifestyle affects human microbiota on daily timescales.Genome Biol. 2016 May 31;17(1):117. doi: 10.1186/s13059-016-0988-y. Genome Biol. 2016. PMID: 27246704 Free PMC article. No abstract available.
Abstract
Background: Disturbance to human microbiota may underlie several pathologies. Yet, we lack a comprehensive understanding of how lifestyle affects the dynamics of human-associated microbial communities.
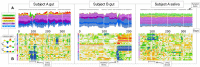
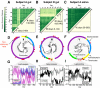
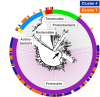
Results: Here, we link over 10,000 longitudinal measurements of human wellness and action to the daily gut and salivary microbiota dynamics of two individuals over the course of one year. These time series show overall microbial communities to be stable for months. However, rare events in each subjects’ life rapidly and broadly impacted microbiota dynamics. Travel from the developed to the developing world in one subject led to a nearly two-fold increase in the Bacteroidetes to Firmicutes ratio, which reversed upon return. Enteric infection in the other subject resulted in the permanent decline of most gut bacterial taxa, which were replaced by genetically similar species. Still, even during periods of overall community stability, the dynamics of select microbial taxa could be associated with specific host behaviors. Most prominently, changes in host fiber intake positively correlated with next-day abundance changes among 15% of gut microbiota members.
Conclusions: Our findings suggest that although human-associated microbial communities are generally stable, they can be quickly and profoundly altered by common human actions and experiences.
Figures

References
-
- Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

