DNA stretching on the wall surfaces in curved microchannels with different radii
- PMID: 25147488
- PMCID: PMC4134334
- DOI: 10.1186/1556-276X-9-382
DNA stretching on the wall surfaces in curved microchannels with different radii
Abstract
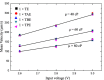
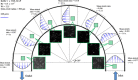
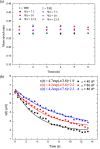
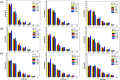
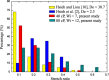
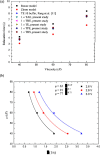
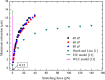
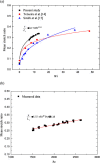
DNA molecule conformation dynamics and stretching were made on semi-circular surfaces with different radii (500 to 5,000 μm) in microchannels measuring 200 μm × 200 μm in cross section. Five different buffer solutions - 1× Tris-acetate-EDTA (TAE), 1× Tris-borate-EDTA (TBE), 1× Tris-EDTA (TE), 1× Tris-phosphate-EDTA (TPE), and 1× Tris-buffered saline (TBS) solutions - were used with a variety of viscosity such as 40, 60, and 80 cP, with resultant 10(-4) ≤ Re ≤ 10(-3) and the corresponding 5 ≤ Wi ≤ 12. The test fluids were seeded with JOJO-1 tracer particles for flow visualization and driven through the test channels via a piezoelectric (PZT) micropump. Micro particle image velocimetry (μPIV) measuring technique was applied for the centered-plane velocity distribution measurements. It is found that the radius effect on the stretch ratio of DNA dependence is significant. The stretch ratio becomes larger as the radius becomes small due to the larger centrifugal force. Consequently, the maximum stretch was found at the center of the channel with a radius of 500 μm.
Keywords: Curve effect; DNA stretching; Microchannel; μPIV.
Figures



References
-
- Randall GC, Schultz KM, Doyle PS. Methods to electrophoretically stretch DNA: microcontractions, gels, and hybrid gel-microcontraction devices. Lab Chip. 2006;9:516–525. - PubMed
-
- Gulati S, Liepmann D, Muller SJ. Elastic secondary flows of semidilute DNA solutions in abrupt 90° microbends. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;9:036314. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

