Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs
- PMID: 25147954
- PMCID: PMC4143927
- DOI: 10.1038/ncomms5684
Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs
Abstract
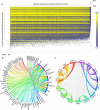
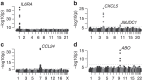
Ideal biomarkers used for disease diagnosis should display deviating levels in affected individuals only and be robust to factors unrelated to the disease. Here we show the impact of genetic, clinical and lifestyle factors on circulating levels of 92 protein biomarkers for cancer and inflammation, using a population-based cohort of 1,005 individuals. For 75% of the biomarkers, the levels are significantly heritable and genome-wide association studies identifies 16 novel loci and replicate 2 previously known loci with strong effects on one or several of the biomarkers with P-values down to 4.4 × 10(-58). Integrative analysis attributes as much as 56.3% of the observed variance to non-disease factors. We propose that information on the biomarker-specific profile of major genetic, clinical and lifestyle factors should be used to establish personalized clinical cutoffs, and that this would increase the sensitivity of using biomarkers for prediction of clinical end points.
Conflict of interest statement
Ulf Gyllensten and Stefan Enroth are authors on a patent application entitled 'Determination and analysis of Biomarkers in clinical samples'; serial number GB1410956.5 (2014). The remaining authors declare no competing financial interests.
Figures

References
-
- Yue C. et al.. The effects of adalimumab and methotrexate treatment on peripheral Th17 cells and IL-17/IL-6 secretion in rheumatoid arthritis patients. Rheumatol. Int. 30, 1553–1557 (2010). - PubMed
-
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
-
- Cao Y., DePinho R. A., Ernst M. & Vousden K. Cancer research: past, present and future. Nat. Rev. Cancer 11, 749–754 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

