Genomic epidemiology of Salmonella enterica serotype Enteritidis based on population structure of prevalent lineages
- PMID: 25147968
- PMCID: PMC4178404
- DOI: 10.3201/eid2009.131095
Genomic epidemiology of Salmonella enterica serotype Enteritidis based on population structure of prevalent lineages
Abstract
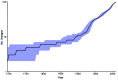
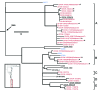
Salmonella enterica serotype Enteritidis is one of the most commonly reported causes of human salmonellosis. Its low genetic diversity, measured by fingerprinting methods, has made subtyping a challenge. We used whole-genome sequencing to characterize 125 S. enterica Enteritidis and 3 S. enterica serotype Nitra strains. Single-nucleotide polymorphisms were filtered to identify 4,887 reliable loci that distinguished all isolates from each other. Our whole-genome single-nucleotide polymorphism typing approach was robust for S. enterica Enteritidis subtyping with combined data for different strains from 2 different sequencing platforms. Five major genetic lineages were recognized, which revealed possible patterns of geographic and epidemiologic distribution. Analyses on the population dynamics and evolutionary history estimated that major lineages emerged during the 17th-18th centuries and diversified during the 1920s and 1950s.
Figures


References
-
- Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. 2011;8:887–900. 10.1089/fpd.2010.0787 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

