In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation
- PMID: 25148027
- PMCID: PMC4160313
- DOI: 10.1016/j.immuni.2014.08.002
In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation
Abstract
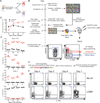
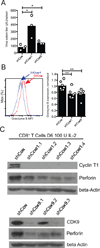
Classical genetic approaches to examine the requirements of genes for T cell differentiation during infection are time consuming. Here we developed a pooled approach to screen 30-100+ genes individually in separate antigen-specific T cells during infection using short hairpin RNAs in a microRNA context (shRNAmir). Independent screens using T cell receptor (TCR)-transgenic CD4(+) and CD8(+) T cells responding to lymphocytic choriomeningitis virus (LCMV) identified multiple genes that regulated development of follicular helper (Tfh) and T helper 1 (Th1) cells, and short-lived effector and memory precursor cytotoxic T lymphocytes (CTLs). Both screens revealed roles for the positive transcription elongation factor (P-TEFb) component Cyclin T1 (Ccnt1). Inhibiting expression of Cyclin T1, or its catalytic partner Cdk9, impaired development of Th1 cells and protective short-lived effector CTL and enhanced Tfh cell and memory precursor CTL formation in vivo. This pooled shRNA screening approach should have utility in numerous immunological studies.
Copyright © 2014 Elsevier Inc. All rights reserved.
Conflict of interest statement
We have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

