A multiple-SNP approach for genome-wide association study of milk production traits in Chinese Holstein cattle
- PMID: 25148050
- PMCID: PMC4141689
- DOI: 10.1371/journal.pone.0099544
A multiple-SNP approach for genome-wide association study of milk production traits in Chinese Holstein cattle
Abstract
The multiple-SNP analysis has been studied by many researchers, in which the effects of multiple SNPs are simultaneously estimated and tested in a multiple linear regression. The multiple-SNP association analysis usually has higher power and lower false-positive rate for detecting causative SNP(s) than single marker analysis (SMA). Several methods have been proposed to simultaneously estimate and test multiple SNP effects. In this research, a fast method called MEML (Mixed model based Expectation-Maximization Lasso algorithm) was developed for simultaneously estimate of multiple SNP effects. An improved Lasso prior was assigned to SNP effects which were estimated by searching the maximum joint posterior mode. The residual polygenic effect was included in the model to absorb many tiny SNP effects, which is treated as missing data in our EM algorithm. A series of simulation experiments were conducted to validate the proposed method, and the results showed that compared with SMMA, the new method can dramatically decrease the false-positive rate. The new method was also applied to the 50k SNP-panel dataset for genome-wide association study of milk production traits in Chinese Holstein cattle. Totally, 39 significant SNPs and their nearby 25 genes were found. The number of significant SNPs is remarkably fewer than that by SMMA which found 105 significant SNPs. Among 39 significant SNPs, 8 were also found by SMMA and several well-known QTLs or genes were confirmed again; furthermore, we also got some positional candidate gene with potential function of effecting milk production traits. These novel findings in our research should be valuable for further investigation.
Conflict of interest statement
Figures
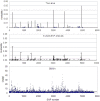
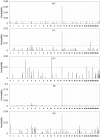
References
-
- Lu B, Zhang D, McCammon JA (2005) Computation of electrostatic forces between solvated molecules determined by the Poisson-Boltzmann equation using a boundary element method. J Chem Phys 122: 214102. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

