Eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay by two genetically separable mechanisms
- PMID: 25148142
- PMCID: PMC4141738
- DOI: 10.1371/journal.pone.0104391
Eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay by two genetically separable mechanisms
Abstract
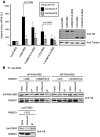
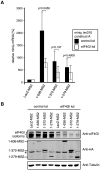
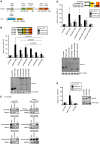
Nonsense-mediated mRNA decay (NMD), which is best known for degrading mRNAs with premature termination codons (PTCs), is thought to be triggered by aberrant translation termination at stop codons located in an environment of the mRNP that is devoid of signals necessary for proper termination. In mammals, the cytoplasmic poly(A)-binding protein 1 (PABPC1) has been reported to promote correct termination and therewith antagonize NMD by interacting with the eukaryotic release factors 1 (eRF1) and 3 (eRF3). Using tethering assays in which proteins of interest are recruited as MS2 fusions to a NMD reporter transcript, we show that the three N-terminal RNA recognition motifs (RRMs) of PABPC1 are sufficient to antagonize NMD, while the eRF3-interacting C-terminal domain is dispensable. The RRM1-3 portion of PABPC1 interacts with eukaryotic initiation factor 4G (eIF4G) and tethering of eIF4G to the NMD reporter also suppresses NMD. We identified the interactions of the eIF4G N-terminus with PABPC1 and the eIF4G core domain with eIF3 as two genetically separable features that independently enable tethered eIF4G to inhibit NMD. Collectively, our results reveal a function of PABPC1, eIF4G and eIF3 in translation termination and NMD suppression, and they provide additional evidence for a tight coupling between translation termination and initiation.
Conflict of interest statement
Figures


References
-
- He F, Li X, Spatrick P, Casillo R, Dong S, et al. (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452. - PubMed
-
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

