Molecular phylogeography and population genetic structure of O. longilobus and O. taihangensis (Opisthopappus) on the Taihang mountains
- PMID: 25148249
- PMCID: PMC4141751
- DOI: 10.1371/journal.pone.0104773
Molecular phylogeography and population genetic structure of O. longilobus and O. taihangensis (Opisthopappus) on the Taihang mountains
Abstract
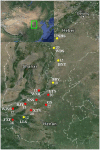
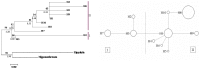
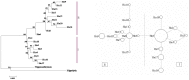
Historic events such as the uplift of mountains and climatic oscillations in the Quaternary periods greatly affected the evolution and modern distribution of the flora. We sequenced the trnL-trnF, ndhJ-trnL and ITS from populations throughout the known distributions of O. longilobus and O. taihangensis to understand the evolutionary history and the divergence related to the past shifts of habitats in the Taihang Mountains regions. The results showed high genetic diversity and pronounced genetic differentiation among the populations of the two species with a significant phylogeographical pattern (NST>GST, P<0.05), which imply restricted gene flow among the populations and significant geographical or environmental isolation. Ten chloroplast DNA (cpDNA) and eighteen nucleus ribosome DNA (nrDNA) haplotypes were identified and clustered into two lineages. Two corresponding refuge areas were revealed across the entire distribution ranges of O. longilobus and at least three refuge areas for O. taihangensis. O. longilobus underwent an evolutionary historical process of long-distance dispersal and colonization, whereas O. taihangensis underwent a population expansion before the main uplift of Taihang Mountains. The differentiation time between O. longilobus and O. taihangensis is estimated to have occurred at the early Pleistocene. Physiographic complexity and paleovegetation transition of Taihang Mountains mainly shaped the specific formation and effected the present distribution of these two species. The results therefore support the inference that Quaternary refugial isolation promoted allopatric speciation in Taihang Mountains. This may help to explain the existence of high diversity and endemism of plant species in central/northern China.
Conflict of interest statement
Figures

Similar articles
-
Localized environmental heterogeneity drives the population differentiation of two endangered and endemic Opisthopappus Shih species.BMC Ecol Evol. 2021 Apr 15;21(1):56. doi: 10.1186/s12862-021-01790-0. BMC Ecol Evol. 2021. PMID: 33858342 Free PMC article.
-
Phylogeographic analysis of a temperate-deciduous forest restricted plant (Bupleurum longiradiatum Turcz.) reveals two refuge areas in China with subsequent refugial isolation promoting speciation.Mol Phylogenet Evol. 2013 Sep;68(3):628-43. doi: 10.1016/j.ympev.2013.04.007. Epub 2013 Apr 23. Mol Phylogenet Evol. 2013. PMID: 23624194
-
High genetic diversity and insignificant interspecific differentiation in Opisthopappus Shih, an endangered cliff genus endemic to the Taihang Mountains of China.ScientificWorldJournal. 2013 Dec 17;2013:275753. doi: 10.1155/2013/275753. eCollection 2013. ScientificWorldJournal. 2013. PMID: 24453824 Free PMC article.
-
Chloroplast DNA phylogeography of Clintoniaudensis Trautv. & Mey. (Liliaceae) in East Asia.Mol Phylogenet Evol. 2010 May;55(2):721-32. doi: 10.1016/j.ympev.2010.02.010. Epub 2010 Feb 19. Mol Phylogenet Evol. 2010. PMID: 20172032 Review.
-
Mountains as Evolutionary Arenas: Patterns, Emerging Approaches, Paradigm Shifts, and Their Implications for Plant Phylogeographic Research in the Tibeto-Himalayan Region.Front Plant Sci. 2019 Mar 18;10:195. doi: 10.3389/fpls.2019.00195. eCollection 2019. Front Plant Sci. 2019. PMID: 30936883 Free PMC article. Review.
Cited by
-
Late Pleistocene speciation of three closely related tree peonies endemic to the Qinling-Daba Mountains, a major glacial refugium in Central China.Ecol Evol. 2019 Jun 17;9(13):7528-7548. doi: 10.1002/ece3.5284. eCollection 2019 Jul. Ecol Evol. 2019. PMID: 31346420 Free PMC article.
-
Adaptation insights from comparative transcriptome analysis of two Opisthopappus species in the Taihang mountains.BMC Genomics. 2022 Jun 24;23(1):466. doi: 10.1186/s12864-022-08703-5. BMC Genomics. 2022. PMID: 35751010 Free PMC article.
-
Localized environmental heterogeneity drives the population differentiation of two endangered and endemic Opisthopappus Shih species.BMC Ecol Evol. 2021 Apr 15;21(1):56. doi: 10.1186/s12862-021-01790-0. BMC Ecol Evol. 2021. PMID: 33858342 Free PMC article.
-
De novo Assembly and Transcriptome Characterization of Opisthopappus (Asteraceae) for Population Differentiation and Adaption.Front Genet. 2018 Sep 19;9:371. doi: 10.3389/fgene.2018.00371. eCollection 2018. Front Genet. 2018. PMID: 30283491 Free PMC article.
-
Root Physiological Traits and Transcriptome Analyses Reveal that Root Zone Water Retention Confers Drought Tolerance to Opisthopappus taihangensis.Sci Rep. 2020 Feb 14;10(1):2627. doi: 10.1038/s41598-020-59399-0. Sci Rep. 2020. PMID: 32060321 Free PMC article.
References
-
- Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. - PubMed
-
- Willis KJ, Whittaker RJ (2002) Species diversity-scale matters. Science 1245–1248. - PubMed
-
- Turchetto-Zolet AC, Cruz F, Vendramin GG, Simon MF, Salgueiro F, et al. (2012) Large-scale phylogeography of the disjunct Neotropical tree species Schizolobium parahyba (Fabaceae-Caesalpinioideae). Molecular Phylogenetics and Evolution 65: 174–182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

