Beyond BRAF(V600): clinical mutation panel testing by next-generation sequencing in advanced melanoma
- PMID: 25148578
- PMCID: PMC4289407
- DOI: 10.1038/jid.2014.366
Beyond BRAF(V600): clinical mutation panel testing by next-generation sequencing in advanced melanoma
Abstract
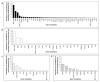
The management of melanoma has evolved owing to improved understanding of its molecular drivers. To augment the current understanding of the prevalence, patterns, and associations of mutations in this disease, the results of clinical testing of 699 advanced melanoma patients using a pan-cancer next-generation sequencing (NGS) panel of hotspot regions in 46 genes were reviewed. Mutations were identified in 43 of the 46 genes on the panel. The most common mutations were BRAFV600 (36%), NRAS (21%), TP53 (16%), BRAFNon-V600 (6%), and KIT (4%). Approximately one-third of melanomas had >1 mutation detected, and the number of mutations per tumor was associated with melanoma subtype. Concurrent TP53 mutations were the most frequent events in tumors with BRAFV600 and NRAS mutations. Melanomas with BRAFNon-V600mutations frequently harbored concurrent NRAS mutations (18%), which were rare in tumors with BRAFV600 mutations (1.6%). The prevalence of BRAFV600 and KIT mutations were significantly associated with melanoma subtypes, and BRAFV600 and TP53 mutations were significantly associated with cutaneous primary tumor location. Multiple potential therapeutic targets were identified in metastatic unknown primary and cutaneous melanomas that lacked BRAFV600 and NRAS mutations. These results enrich our understanding of the patterns and clinical associations of oncogenic mutations in melanoma.
Conflict of interest statement
Figures



Comment in
-
Panel sequencing melanomas.J Invest Dermatol. 2015 Feb;135(2):335-336. doi: 10.1038/jid.2014.420. J Invest Dermatol. 2015. PMID: 25573045
References
-
- Albino AP, Vidal MJ, McNutt NS, et al. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 1994;4:35–45. - PubMed
-
- Bucheit AD, Syklawer E, Jakob JA, et al. Clinical characteristics and outcomes with specific BRAF and NRAS mutations in patients with metastatic melanoma. Cancer. 2013;119:3821–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

