Structure-based virtual screening of the nociceptin receptor: hybrid docking and shape-based approaches for improved hit identification
- PMID: 25148595
- PMCID: PMC4210177
- DOI: 10.1021/ci500291a
Structure-based virtual screening of the nociceptin receptor: hybrid docking and shape-based approaches for improved hit identification
Abstract
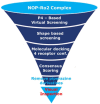
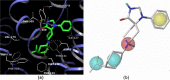
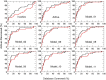
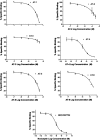
The antagonist-bound crystal structure of the nociceptin receptor (NOP), from the opioid receptor family, was recently reported along with those of the other opioid receptors bound to opioid antagonists. We recently reported the first homology model of the 'active-state' of the NOP receptor, which when docked with 'agonist' ligands showed differences in the TM helices and residues, consistent with GPCR activation after agonist binding. In this study, we explored the use of the active-state NOP homology model for structure-based virtual screening to discover NOP ligands containing new chemical scaffolds. Several NOP agonist and antagonist ligands previously reported are based on a common piperidine scaffold. Given the structure-activity relationships for known NOP ligands, we developed a hybrid method that combines a structure-based and ligand-based approach, utilizing the active-state NOP receptor as well as the pharmacophoric features of known NOP ligands, to identify novel NOP binding scaffolds by virtual screening. Multiple conformations of the NOP active site including the flexible second extracellular loop (EL2) loop were generated by simulated annealing and ranked using enrichment factor (EF) analysis and a ligand-decoy dataset containing known NOP agonist ligands. The enrichment factors were further improved by combining shape-based screening of this ligand-decoy dataset and calculation of consensus scores. This combined structure-based and ligand-based EF analysis yielded higher enrichment factors than the individual methods, suggesting the effectiveness of the hybrid approach. Virtual screening of the CNS Permeable subset of the ZINC database was carried out using the above-mentioned hybrid approach in a tiered fashion utilizing a ligand pharmacophore-based filtering step, followed by structure-based virtual screening using the refined NOP active-state models from the enrichment analysis. Determination of the NOP receptor binding affinity of a selected set of top-scoring hits resulted in identification of several compounds with measurable binding affinity at the NOP receptor, one of which had a new chemotype for NOP receptor binding. The hybrid ligand-based and structure-based methodology demonstrates an effective approach for virtual screening that leverages existing SAR and receptor structure information for identifying novel hits for NOP receptor binding. The refined active-state NOP homology models obtained from the enrichment studies can be further used for structure-based optimization of these new chemotypes to obtain potent and selective NOP receptor ligands for therapeutic development.
Figures




Similar articles
-
Homology modeling and molecular dynamics simulations of the active state of the nociceptin receptor reveal new insights into agonist binding and activation.Proteins. 2012 Aug;80(8):1948-61. doi: 10.1002/prot.24077. Epub 2012 May 17. Proteins. 2012. PMID: 22489047 Free PMC article.
-
Discovery of the first small-molecule opioid pan antagonist with nanomolar affinity at mu, delta, kappa, and nociceptin opioid receptors.ACS Chem Neurosci. 2015 Apr 15;6(4):646-57. doi: 10.1021/cn500367b. Epub 2015 Feb 18. ACS Chem Neurosci. 2015. PMID: 25635572 Free PMC article.
-
Small-molecule agonists and antagonists of the opioid receptor-like receptor (ORL1, NOP): ligand-based analysis of structural factors influencing intrinsic activity at NOP.AAPS J. 2005 Oct 5;7(2):E345-52. doi: 10.1208/aapsj070234. AAPS J. 2005. PMID: 16353914 Free PMC article. Review.
-
"In silico" study of the binding of two novel antagonists to the nociceptin receptor.J Comput Aided Mol Des. 2018 Feb;32(2):385-400. doi: 10.1007/s10822-017-0095-5. Epub 2018 Jan 16. J Comput Aided Mol Des. 2018. PMID: 29340866
-
Synthesis and biological activity of small peptides as NOP and opioid receptors' ligands: view on current developments.Vitam Horm. 2015;97:123-46. doi: 10.1016/bs.vh.2014.11.005. Epub 2015 Jan 22. Vitam Horm. 2015. PMID: 25677770 Review.
Cited by
-
Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design.Front Pharmacol. 2018 Mar 9;9:128. doi: 10.3389/fphar.2018.00128. eCollection 2018. Front Pharmacol. 2018. PMID: 29593527 Free PMC article. Review.
-
Proposed Mode of Binding and Action of Positive Allosteric Modulators at Opioid Receptors.ACS Chem Biol. 2016 May 20;11(5):1220-9. doi: 10.1021/acschembio.5b00712. Epub 2016 Feb 17. ACS Chem Biol. 2016. PMID: 26841170 Free PMC article.
-
The Use of Computational Approaches in the Discovery and Mechanism Study of Opioid Analgesics.Front Chem. 2020 May 15;8:335. doi: 10.3389/fchem.2020.00335. eCollection 2020. Front Chem. 2020. PMID: 32500054 Free PMC article. Review.
-
Fusing Docking Scoring Functions Improves the Virtual Screening Performance for Discovering Parkinson's Disease Dual Target Ligands.Curr Neuropharmacol. 2017 Nov 14;15(8):1107-1116. doi: 10.2174/1570159X15666170109143757. Curr Neuropharmacol. 2017. PMID: 28067172 Free PMC article.
-
Antinociceptive Activity of Chemical Components of Essential Oils That Involves Docking Studies: A Review.Front Pharmacol. 2020 May 29;11:777. doi: 10.3389/fphar.2020.00777. eCollection 2020. Front Pharmacol. 2020. PMID: 32547391 Free PMC article.
References
-
- Chen Y.; Fan Y.; Liu J.; Mestek A.; Tian M.; Kozak C. A.; Yu L. Molecular cloning, tissue distribution and chromosomal localization of a novel member of the opioid receptor gene family. FEBS Lett. 1994, 347, 279–83. - PubMed
-
- Bunzow J. R.; Saez C.; Mortrud M.; Bouvier C.; Williams J. T.; Low M.; Grandy D. K. Molecular cloning and tissue distribution of a putative member of the rat opioid receptor gene family that is not a mu, delta or kappa opioid receptor type. FEBS Lett. 1994, 347, 284–8. - PubMed
-
- Mollereau C.; Parmentier M.; Mailleux P.; Butour J. L.; Moisand C.; Chalon P.; Caput D.; Vassart G.; Meunier J. C. ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS Lett. 1994, 341, 33–8. - PubMed
-
- Bignan G. C.; Connolly P. J.; Middleton S. A. Recent advances towards the discovery of ORL-1 receptor agonists and antagonists. Expert Opin. Ther. Pat. 2005, 15, 357–388.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

