Newly identified thermostable esterase from Sulfobacillus acidophilus: properties and performance in phthalate ester degradation
- PMID: 25149523
- PMCID: PMC4249001
- DOI: 10.1128/AEM.02072-14
Newly identified thermostable esterase from Sulfobacillus acidophilus: properties and performance in phthalate ester degradation
Abstract
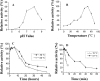
EstS1, a newly identified thermostable esterase from Sulfobacillus acidophilus DSM10332, was heterologously expressed in Escherichia coli and shown to enzymatically degrade phthalate esters (PAEs) to their corresponding monoalkyl PAEs. The optimal pH and temperature of the esterase were found to be 8.0 and 70°C, respectively. The half-life of EstS1 at 60°C was 15 h, indicating that the enzyme had good thermostability. The specificity constant (kcat/Km) of the enzyme for p-nitrophenyl butyrate was as high as 6,770 mM(-1) s(-1). The potential value of EstS1 was demonstrated by its ability to effectively hydrolyze 35 to 82% of PAEs (10 mM) within 2 min at 37°C, with all substrates being completely degraded within 24 h. At 60°C, the time required for complete hydrolysis of most PAEs was reduced by half. To our knowledge, this enzyme is a new esterase identified from thermophiles that is able to degrade various PAEs at high temperatures.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Gu JD, Li J, Wang Y. 2005. Biochemical pathway and degradation of phthalate ester isomers by bacteria. Water Sci. Technol. 52:241–248. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

