FusoBase: an online Fusobacterium comparative genomic analysis platform
- PMID: 25149689
- PMCID: PMC4141642
- DOI: 10.1093/database/bau082
FusoBase: an online Fusobacterium comparative genomic analysis platform
Abstract
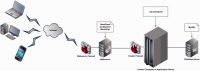
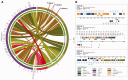
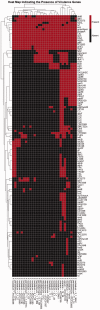
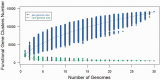
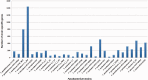
Fusobacterium are anaerobic gram-negative bacteria that have been associated with a wide spectrum of human infections and diseases. As the biology of Fusobacterium is still not well understood, comparative genomic analysis on members of this species will provide further insights on their taxonomy, phylogeny, pathogenicity and other information that may contribute to better management of infections and diseases. To facilitate the ongoing genomic research on Fusobacterium, a specialized database with easy-to-use analysis tools is necessary. Here we present FusoBase, an online database providing access to genome-wide annotated sequences of Fusobacterium strains as well as bioinformatics tools, to support the expanding scientific community. Using our custom-developed Pairwise Genome Comparison tool, we demonstrate how differences between two user-defined genomes and how insertion of putative prophages can be identified. In addition, Pathogenomics Profiling Tool is capable of clustering predicted genes across Fusobacterium strains and visualizing the results in the form of a heat map with dendrogram.
Database url: http://fusobacterium.um.edu.my.
© The Author(s) 2014. Published by Oxford University Press.
Figures


Similar articles
-
YersiniaBase: a genomic resource and analysis platform for comparative analysis of Yersinia.BMC Bioinformatics. 2015 Jan 16;16(1):9. doi: 10.1186/s12859-014-0422-y. BMC Bioinformatics. 2015. PMID: 25591325 Free PMC article.
-
CoryneBase: Corynebacterium genomic resources and analysis tools at your fingertips.PLoS One. 2014 Jan 17;9(1):e86318. doi: 10.1371/journal.pone.0086318. eCollection 2014. PLoS One. 2014. PMID: 24466021 Free PMC article.
-
StaphyloBase: a specialized genomic resource for the staphylococcal research community.Database (Oxford). 2014 Feb 26;2014:bau010. doi: 10.1093/database/bau010. Print 2014. Database (Oxford). 2014. PMID: 24578355 Free PMC article.
-
Assembly, Annotation, and Comparative Genomics in PATRIC, the All Bacterial Bioinformatics Resource Center.Methods Mol Biol. 2018;1704:79-101. doi: 10.1007/978-1-4939-7463-4_4. Methods Mol Biol. 2018. PMID: 29277864 Review.
-
Integration of bioinformatics to biodegradation.Biol Proced Online. 2014 Apr 27;16:8. doi: 10.1186/1480-9222-16-8. eCollection 2014. Biol Proced Online. 2014. PMID: 24808763 Free PMC article. Review.
Cited by
-
Prevention and Control of Pathogens Based on Big-Data Mining and Visualization Analysis.Front Mol Biosci. 2021 Feb 25;7:626595. doi: 10.3389/fmolb.2020.626595. eCollection 2020. Front Mol Biosci. 2021. PMID: 33718431 Free PMC article.
-
Characterization of the non-glandular gastric region microbiota in Helicobacter suis-infected versus non-infected pigs identifies a potential role for Fusobacterium gastrosuis in gastric ulceration.Vet Res. 2019 May 24;50(1):39. doi: 10.1186/s13567-019-0656-9. Vet Res. 2019. PMID: 31126330 Free PMC article.
-
Noma (cancrum oris): An unresolved global challenge.Periodontol 2000. 2019 Jun;80(1):189-199. doi: 10.1111/prd.12275. Periodontol 2000. 2019. PMID: 31090145 Free PMC article. Review.
References
-
- Citron D.M., Poxton I.R., Baron E.J. (2007) Bacteroides, porphyromonas, prevotella, fusobacterium, and other anaerobic gram-negative rods. In: Murray P.R., Baron E.J., Landry M.L., et al.. (eds), Manual of Clinical Microbiology. ASM Press, Washington, DC, pp. 911–932
-
- Strauss J., Kaplan G.G., Beck P.L., et al. (2011) Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis., 17, 1971–1978 - PubMed
-
- Gonzales-Marin C., Spratt D.A., Allaker R.P. (2013) Maternal oral origin of Fusobacterium nucleatum in adverse pregnancy outcomes as determined using the 16S-23S rRNA gene intergenic transcribed spacer region. J. Med. Microbiol., 62, 133–144 - PubMed
-
- Huggan P.J., Murdoch D.R. (2008) Fusobacterial infections: clinical spectrum and incidence of invasive disease. J. Infect., 57, 283–289 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

