A renewal theory approach to IBD sharing
- PMID: 25149691
- PMCID: PMC4179929
- DOI: 10.1016/j.tpb.2014.08.002
A renewal theory approach to IBD sharing
Abstract
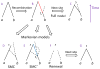
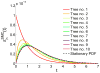
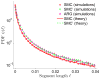
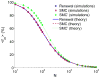
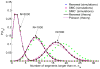
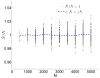
A long genomic segment inherited by a pair of individuals from a single, recent common ancestor is said to be identical-by-descent (IBD). Shared IBD segments have numerous applications in genetics, from demographic inference to phasing, imputation, pedigree reconstruction, and disease mapping. Here, we provide a theoretical analysis of IBD sharing under Markovian approximations of the coalescent with recombination. We describe a general framework for the IBD process along the chromosome under the Markovian models (SMC/SMC'), as well as introduce and justify a new model, which we term the renewal approximation, under which lengths of successive segments are independent. Then, considering the infinite-chromosome limit of the IBD process, we recover previous results (for SMC) and derive new results (for SMC') for the mean number of shared segments longer than a cutoff and the fraction of the chromosome found in such segments. We then use renewal theory to derive an expression (in Laplace space) for the distribution of the number of shared segments and demonstrate implications for demographic inference. We also compute (again, in Laplace space) the distribution of the fraction of the chromosome in shared segments, from which we obtain explicit expressions for the first two moments. Finally, we generalize all results to populations with a variable effective size.
Keywords: Coalescent theory; IBD sharing; Recombination; Renewal theory; SMC; SMC’.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, Ostrer H. Abraham’s children in the genome era: Major Jewish diaspora populations comprise distinct genetic clusters with shared middle eastern ancestry. Am J Hum Genet. 2010;86:850–859. - PMC - PubMed
-
- Botigué LR, Henn BM, Gravel S, Maples BK, Gignoux CR, Corona E, Atzmon G, Burns E, Ostrer H, Flores C, Bertranpetit J, Comas D, Bustamante CD. Gene flow from North Africa contributes to differential human genetic diversity in Southern Europe. Proc Natl Acad Sci USA. 2013;110:11791–11796. - PMC - PubMed
-
- Brančík L. Numerical Inverse Laplace Transforms for Electrical Engineering Simulation, MATLAB for Engineers - Applications in Control, Electrical Engineering, IT and Robotics. InTech; 2011.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

