Regulation of fibroblast lipid storage and myofibroblast phenotypes during alveolar septation in mice
- PMID: 25150063
- PMCID: PMC4200388
- DOI: 10.1152/ajplung.00144.2014
Regulation of fibroblast lipid storage and myofibroblast phenotypes during alveolar septation in mice
Abstract
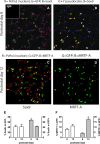
Signaling through platelet-derived growth factor receptor-α (PDGFRα) is required for alveolar septation and participates in alveolar regeneration after pneumonectomy. In both adipose tissue and skeletal muscle, bipotent pdgfrα-expressing progenitors expressing delta-like ligand-1 or sex-determining region Y box 9 (Sox9) may differentiate into either lipid storage cells or myofibroblasts. We analyzed markers of mesenchymal progenitors and differentiation in lung fibroblasts (LF) with different levels (absent, low, or high) of pdgfrα gene expression. A larger proportion of pdgfrα-expressing than nonexpressing LF contained Sox9. Neutral lipids, CD166, and Tcf21 were more abundant in LF with a lower compared with a higher level of pdgfrα gene expression. PDGF-A increased Sox9 in primary LF cultures, suggesting that active signaling through PDGFRα is required to maintain Sox9. As alveolar septation progresses from postnatal day (P) 8 to P12, fewer pdgfrα-expressing LF contain Sox9, whereas more of these LF contain myocardin-like transcription factor-A, showing that Sox9 diminishes as LF become myofibroblasts. At P8, neutral lipid droplets predominate in LF with the lower level of pdgfrα gene expression, whereas transgelin (tagln) was predominantly expressed in LF with higher pdgfrα gene expression. Targeted deletion of pdgfrα in LF, which expressed tagln, reduced Sox9 in α-actin (α-SMA, ACTA2)-containing LF, whereas it increased the abundance of cell surface delta-like protein-1 (as well as peroxisome proliferator-activated receptor-γ and tcf21 mRNA in LF, which also expressed stem cell antigen-1). Thus pdgfrα deletion differentially alters delta-like protein-1 and Sox9, suggesting that targeting different downstream pathways in PDGF-A-responsive LF could identify strategies that promote lung regeneration without initiating fibrosis.
Keywords: adipocyte; delta-like ligand-1 (Dlk1); lung alveolarization; platelet-derived growth factor; preadipocyte factor-1; sex-determining region Y box 9; stem cell; stereology.
Figures









References
-
- Andersen DC, Laborda J, Baladron V, Kassem M, Sheikh SP, Jensen CH. Dual role of delta-like 1 homolog (DLK1) in skeletal muscle development and adult muscle regeneration. Development 140: 3743–3753, 2013 - PubMed
-
- Boström H, Gritli-Linde A, Betsholtz C. PDGF-A/PDGF alpha-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis. Dev Dynam 223: 155–162, 2002 - PubMed
-
- Boström H, Willetts K, Pekny M, Leveen P, Lindahl P, Hedstrand H, Pekna M, Hellstrom M, Gebre-Medin S, Schalling M, Nilsson M, Kurland S, Tornell J, Heath JK, Betsholtz C. PGDF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85: 863–873, 1996 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

