The G protein α subunit Gαs is a tumor suppressor in Sonic hedgehog-driven medulloblastoma
- PMID: 25150496
- PMCID: PMC4334261
- DOI: 10.1038/nm.3666
The G protein α subunit Gαs is a tumor suppressor in Sonic hedgehog-driven medulloblastoma
Abstract
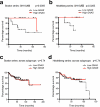
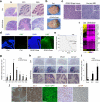
Medulloblastoma, the most common malignant childhood brain tumor, exhibits distinct molecular subtypes and cellular origins. Genetic alterations driving medulloblastoma initiation and progression remain poorly understood. Herein, we identify GNAS, encoding the G protein Gαs, as a potent tumor suppressor gene that, when expressed at low levels, defines a subset of aggressive Sonic hedgehog (SHH)-driven human medulloblastomas. Ablation of the single Gnas gene in anatomically distinct progenitors in mice is sufficient to induce Shh-associated medulloblastomas, which recapitulate their human counterparts. Gαs is highly enriched at the primary cilium of granule neuron precursors and suppresses Shh signaling by regulating both the cAMP-dependent pathway and ciliary trafficking of Hedgehog pathway components. Elevation in levels of a Gαs effector, cAMP, effectively inhibits tumor cell proliferation and progression in Gnas-ablated mice. Thus, our gain- and loss-of-function studies identify a previously unrecognized tumor suppressor function for Gαs that can be found consistently across Shh-group medulloblastomas of disparate cellular and anatomical origins, highlighting G protein modulation as a potential therapeutic avenue.
Figures




References
-
- Grammel D, et al. Sonic hedgehog-associated medulloblastoma arising from the cochlear nuclei of the brainstem. Acta neuropathologica. 2012;123:601–614. doi:10.1007/s00401-012-0961-0. - PubMed
-
- Sasai K, et al. Medulloblastomas derived from Cxcr6 mutant mice respond to treatment with a smoothened inhibitor. Cancer research. 2007;67:3871–3877. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

