Detecting and correcting systematic variation in large-scale RNA sequencing data
- PMID: 25150837
- PMCID: PMC4160374
- DOI: 10.1038/nbt.3000
Detecting and correcting systematic variation in large-scale RNA sequencing data
Abstract
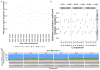
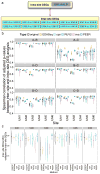
High-throughput RNA sequencing (RNA-seq) enables comprehensive scans of entire transcriptomes, but best practices for analyzing RNA-seq data have not been fully defined, particularly for data collected with multiple sequencing platforms or at multiple sites. Here we used standardized RNA samples with built-in controls to examine sources of error in large-scale RNA-seq studies and their impact on the detection of differentially expressed genes (DEGs). Analysis of variations in guanine-cytosine content, gene coverage, sequencing error rate and insert size allowed identification of decreased reproducibility across sites. Moreover, commonly used methods for normalization (cqn, EDASeq, RUV2, sva, PEER) varied in their ability to remove these systematic biases, depending on sample complexity and initial data quality. Normalization methods that combine data from genes across sites are strongly recommended to identify and remove site-specific effects and can substantially improve RNA-seq studies.
Figures



References
-
- Irizarry RA, et al. Multiple-laboratory comparison of microarray platforms. Nature methods. 2005;2:345–350. - PubMed
-
- Casciano DA, Woodcock J. Empowering microarrays in the regulatory setting. Nature biotechnology. 2006;24:1103. - PubMed
-
- Ball CA, Brazma A. MGED standards: work in progres. Omics : a journal of integrative biology. 2006;10:138–144. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

