Species determination of Culicoides biting midges via peptide profiling using matrix-assisted laser desorption ionization mass spectrometry
- PMID: 25152308
- PMCID: PMC4158057
- DOI: 10.1186/1756-3305-7-392
Species determination of Culicoides biting midges via peptide profiling using matrix-assisted laser desorption ionization mass spectrometry
Abstract
Background: Culicoides biting midges are vectors of bluetongue and Schmallenberg viruses that inflict large-scale disease epidemics in ruminant livestock in Europe. Methods based on morphological characteristics and sequencing of genetic markers are most commonly employed to differentiate Culicoides to species level. Proteomic methods, however, are also increasingly being used as an alternative method of identification. These techniques have the potential to be rapid and may also offer advantages over DNA-based techniques. The aim of this proof-of-principle study was to develop a simple MALDI-MS based method to differentiate Culicoides from different species by peptide patterns with the additional option of identifying discriminating peptides.
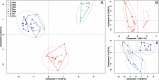
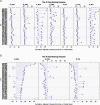
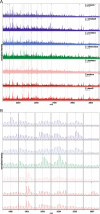
Methods: Proteins extracted from 7 Culicoides species were digested and resulting peptides purified. Peptide mass fingerprint (PMF) spectra were recorded using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) and peak patterns analysed in R using the MALDIquant R package. Additionally, offline liquid chromatography (LC) MALDI-TOF tandem mass spectrometry (MS/MS) was applied to determine the identity of peptide peaks in one exemplary MALDI spectrum obtained using an unfractionated extract.
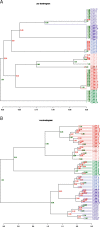
Results: We showed that the majority of Culicoides species yielded reproducible mass spectra with peak patterns that were suitable for classification. The dendrogram obtained by MS showed tentative similarities to a dendrogram generated from cytochrome oxidase I (COX1) sequences. Using offline LC-MALDI-TOF-MS/MS we determined the identity of 28 peptide peaks observed in one MALDI spectrum in a mass range from 1.1 to 3.1 kDa. All identified peptides were identical to other dipteran species and derived from one of five highly abundant proteins due to an absence of available Culicoides data.
Conclusion: Shotgun mass mapping by MALDI-TOF-MS has been shown to be compatible with morphological and genetic identification of specimens. Furthermore, the method performs at least as well as an alternative approach based on MS spectra of intact proteins, thus establishing the procedure as a method in its own right, with the additional option of concurrently using the same samples in other MS-based applications for protein identifications. The future availability of genomic information for different Culicoides species may enable a more stringent peptide detection based on Culicoides-specific sequence information.
Figures


Similar articles
-
Comparison of matrix-assisted laser desorption ionization-time of flight mass spectrometry and molecular biology techniques for identification of Culicoides (Diptera: ceratopogonidae) biting midges in senegal.J Clin Microbiol. 2015 Feb;53(2):410-8. doi: 10.1128/JCM.01855-14. Epub 2014 Nov 19. J Clin Microbiol. 2015. PMID: 25411169 Free PMC article.
-
Identification of field-caught Culicoides biting midges using matrix-assisted laser desorption/ionization time of flight mass spectrometry.Parasitology. 2012 Feb;139(2):248-58. doi: 10.1017/S0031182011001764. Epub 2011 Oct 19. Parasitology. 2012. PMID: 22008297
-
Spatio-temporal occurrence of Culicoides biting midges in the climatic regions of Switzerland, along with large scale species identification by MALDI-TOF mass spectrometry.Parasit Vectors. 2012 Oct 31;5:246. doi: 10.1186/1756-3305-5-246. Parasit Vectors. 2012. PMID: 23111100 Free PMC article.
-
Food allergen analysis: A review of current gaps and the potential to fill them by matrix-assisted laser desorption/ionization.Compr Rev Food Sci Food Saf. 2023 Sep;22(5):3984-4003. doi: 10.1111/1541-4337.13216. Epub 2023 Aug 2. Compr Rev Food Sci Food Saf. 2023. PMID: 37530543 Review.
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry.Clin Chim Acta. 2003 Nov;337(1-2):11-21. doi: 10.1016/j.cccn.2003.08.008. Clin Chim Acta. 2003. PMID: 14568176 Review.
Cited by
-
MALDI-TOF MS Profiling-Advances in Species Identification of Pests, Parasites, and Vectors.Front Cell Infect Microbiol. 2017 May 15;7:184. doi: 10.3389/fcimb.2017.00184. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28555175 Free PMC article. Review.
-
Assessment of MALDI-TOF MS biotyping for Borrelia burgdorferi sl detection in Ixodes ricinus.PLoS One. 2017 Sep 26;12(9):e0185430. doi: 10.1371/journal.pone.0185430. eCollection 2017. PLoS One. 2017. PMID: 28950023 Free PMC article.
-
Assessment of MALDI-TOF mass spectrometry for filariae detection in Aedes aegypti mosquitoes.PLoS Negl Trop Dis. 2017 Dec 20;11(12):e0006093. doi: 10.1371/journal.pntd.0006093. eCollection 2017 Dec. PLoS Negl Trop Dis. 2017. PMID: 29261659 Free PMC article.
-
Culicoides Biting Midges-Underestimated Vectors for Arboviruses of Public Health and Veterinary Importance.Viruses. 2019 Apr 24;11(4):376. doi: 10.3390/v11040376. Viruses. 2019. PMID: 31022868 Free PMC article. Review.
-
MALDI-TOF MS Profiling and Its Contribution to Mosquito-Borne Diseases: A Systematic Review.Insects. 2024 Aug 29;15(9):651. doi: 10.3390/insects15090651. Insects. 2024. PMID: 39336619 Free PMC article. Review.
References
-
- Hoffmann B, Bauer B, Bauer C, Batza HJ, Beer M, Clausen PH, Geier M, Gethmann JM, Kiel E, Liebisch G, Liebisch A, Mehlhorn H, Schaub GA, Werner D, Conraths FJ. Monitoring of putative vectors of bluetongue virus serotype 8, Germany. Emerg Infect Dis. 2009;15:1481–1484. doi: 10.3201/eid1509.090562. - DOI - PMC - PubMed
-
- Elbers AR, Loeffen WL, Quak S, de Boer-Luijtze E, van der Spek AN, Bouwstra R, Maas R, Spierenburg MA, de Kluijver EP, van Schaik G, van der Poel WH. Seroprevalence of Schmallenberg virus antibodies among dairy cattle, the Netherlands, winter 2011–2012. Emerg Infect Dis. 2012;18:1065–1071. doi: 10.3201/eid1807.120323. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

