Functional and genomic context in pathway analysis of GWAS data
- PMID: 25154796
- PMCID: PMC4266582
- DOI: 10.1016/j.tig.2014.07.004
Functional and genomic context in pathway analysis of GWAS data
Abstract
Gene set analysis (GSA) is a promising tool for uncovering the polygenic effects associated with complex diseases. However, the available techniques reflect a wide variety of hypotheses about how genetic effects interact to contribute to disease susceptibility. The lack of consensus about the best way to perform GSA has led to confusion in the field and has made it difficult to compare results across methods. A clear understanding of the various choices made during GSA - such as how gene sets are defined, how single-nucleotide polymorphisms (SNPs) are assigned to genes, and how individual SNP-level effects are aggregated to produce gene- or pathway-level effects - will improve the interpretability and comparability of results across methods and studies. In this review we provide an overview of the various data sources used to construct gene sets and the statistical methods used to test for gene set association, as well as provide guidelines for ensuring the comparability of results.
Keywords: GWAS; complex traits; gene set analysis; polygenic effects.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
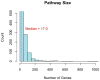
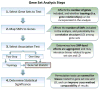
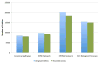

Comment in
-
Pitfalls in the application of gene-set analysis to genetics studies.Trends Genet. 2014 Dec;30(12):513-4. doi: 10.1016/j.tig.2014.10.001. Trends Genet. 2014. PMID: 25459301 Free PMC article.
-
‘Pitfalls in the application of gene set analysis to genetics studies’: a response.Trends Genet. 2014 Dec;30(12):514-5. doi: 10.1016/j.tig.2014.10.002. Trends Genet. 2014. PMID: 25459302 Free PMC article. No abstract available.
References
-
- Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA European Bioinformatics Institute. [Accessed February 19, 2014];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies.
-
- Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009 Apr 23;360(17):1699–701. - PubMed
-
- Holmans P. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Adv Genet. 2010;72:141–79. - PubMed
-
- Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010 Dec;11(12):843–54. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

