Pim1 kinase is upregulated in glioblastoma multiforme and mediates tumor cell survival
- PMID: 25155357
- PMCID: PMC4288523
- DOI: 10.1093/neuonc/nou216
Pim1 kinase is upregulated in glioblastoma multiforme and mediates tumor cell survival
Abstract
Background: The current therapy for glioblastoma multiforme (GBM), the most aggressive and common primary brain tumor of adults, involves surgery and a combined radiochemotherapy that controls tumor progression only for a limited time window. Therefore, the identification of new molecular targets is highly necessary. Inhibition of kinases has become a standard of clinical oncology, and thus the oncogenic kinase Pim1 might represent a promising target for improvement of GBM therapy.
Methods: Expression of Pim1 and associated signaling molecules was analyzed in human GBM samples, and the potential role of this kinase in patients' prognosis was evaluated. Furthermore, we analyzed the in vivo role of Pim1 in GBM cell growth in an orthotopic mouse model and examined the consequences of Pim1 inhibition in vitro to clarify underlying pathways.
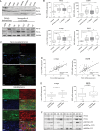
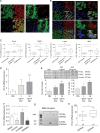
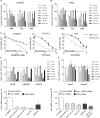
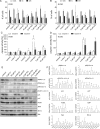
Results: In comparison with normal brain, a strong upregulation of Pim1 was demonstrated in human GBM samples. Notably, patients with short overall survival showed a significantly higher Pim1 expression compared with GBM patients who lived longer than the median. In vitro experiments with GBM cells and analysis of patients' GBM samples suggest that Pim1 regulation is dependent on epidermal growth factor receptor. Furthermore, inhibition of Pim1 resulted in reduced cell viability accompanied by decreased cell numbers and increased apoptotic cells, as seen by elevated subG1 cell contents and caspase-3 and -9 activation, as well as modulation of several cell cycle or apoptosis regulatory proteins.
Conclusions: Altogether, Pim1 could be a novel therapeutic target, which should be further analyzed to improve the outcome of patients with aggressive GBM.
Keywords: epidermal growth factor receptor; glioblastoma multiforme; pim1 kinase.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Ohgaki H, Kleihues P. Genetic profile of astrocytic and oligodendroglial gliomas. Brain Tumor Pathol. 2011;28(3):177–183. - PubMed
-
- Newton HB. Molecular neuro-oncology and development of targeted therapeutic strategies for brain tumors. Part 2: PI3K/Akt/PTEN, mTOR, SHH/PTCH and angiogenesis. Expert Rev Anticancer Ther. 2004;4(1):105–128. - PubMed
-
- Anton K, Baehring JM, Mayer T. Glioblastoma multiforme: overview of current treatment and future perspectives. Hematol Oncol Clin North Am. 2012;26(4):825–853. - PubMed
-
- Cuypers HT, Selten G, Quint W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–150. - PubMed
-
- van Lohuizen M, Verbeek S, Krimpenfort P, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56(4):673–682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

