The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites
- PMID: 25155612
- PMCID: PMC4428586
- DOI: 10.1016/j.molcel.2014.07.012
The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites
Abstract
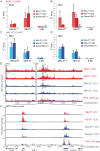
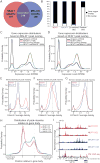
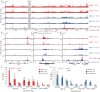
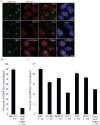
Mechanistic roles for many lncRNAs are poorly understood, in part because their direct interactions with genomic loci and proteins are difficult to assess. Using a method to purify endogenous RNAs and their associated factors, we mapped the genomic binding sites for two highly expressed human lncRNAs, NEAT1 and MALAT1. We show that NEAT1 and MALAT1 localize to hundreds of genomic sites in human cells, primarily over active genes. NEAT1 and MALAT1 exhibit colocalization to many of these loci, but display distinct gene body binding patterns at these sites, suggesting independent but complementary functions for these RNAs. We also identified numerous proteins enriched by both lncRNAs, supporting complementary binding and function, in addition to unique associated proteins. Transcriptional inhibition or stimulation alters localization of NEAT1 on active chromatin sites, implying that underlying DNA sequence does not target NEAT1 to chromatin, and that localization responds to cues involved in the transcription process.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. - PubMed
-
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. - PubMed
-
- Cheung E, Kraus WL. Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol. 2010;72:191–218. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

