Spatial and temporal organization of signaling pathways
- PMID: 25155749
- PMCID: PMC4477539
- DOI: 10.1016/j.tibs.2014.07.008
Spatial and temporal organization of signaling pathways
Abstract
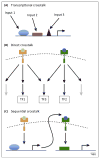
The development and maintenance of the many different cell types in metazoan organisms requires robust and diverse intercellular communication mechanisms. Relatively few such signaling pathways have been identified, leading to the question of how such a broad diversity of output is generated from relatively simple signals. Recent studies have revealed complex mechanisms integrating temporal and spatial information to generate diversity in signaling pathway output. We review some general principles of signaling pathways, focusing on transcriptional outputs in Drosophila. We consider the role of spatial and temporal aspects of different transduction pathways and then discuss how recently developed tools and approaches are helping to dissect the complex mechanisms linking pathway stimulation to output.
Keywords: crosstalk; signaling dynamics; signaling pathways.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

