Transcriptome-wide analysis of messenger RNA decay in normal and osteoarthritic human articular chondrocytes
- PMID: 25155964
- PMCID: PMC4233952
- DOI: 10.1002/art.38849
Transcriptome-wide analysis of messenger RNA decay in normal and osteoarthritic human articular chondrocytes
Abstract
Objective: Messenger RNA (mRNA) decay rates control not only gene expression levels, but also responsiveness to altered transcriptional input. We undertook this study to examine transcriptome-wide posttranscriptional regulation in both normal and osteoarthritic (OA) human articular chondrocytes.
Methods: Human articular chondrocytes were isolated from normal or OA tissue. Equine articular chondrocytes were isolated from young or old horses at a commercial abattoir. RNA decay was measured across the transcriptome in human cells by microarray analysis following an actinomycin D chase. Messenger RNA levels in samples were confirmed using quantitative reverse transcription-polymerase chain reaction.
Results: Examination of total mRNA expression levels demonstrated significant differences in the expression of transcripts between normal and OA chondrocytes. Interestingly, almost no difference was observed in total mRNA expression between chondrocytes from intact OA cartilage and those from fibrillated OA cartilage. Decay analysis revealed a set of rapidly turned over transcripts associated with transcriptional control and programmed cell death that were common to all chondrocytes and contained binding sites for abundant cartilage microRNAs. Many transcripts exhibited altered mRNA half-lives in human OA chondrocytes compared to normal cells. Specific transcripts whose decay rates were altered were generally less stable in these pathologic cells. Examination of selected genes in chondrocytes from young and old healthy horses did not identify any change in mRNA turnover.
Conclusion: This is the first investigation into the "posttranscriptome" of the chondrocyte. It identifies a set of short-lived chondrocyte mRNAs likely to be highly responsive to altered transcriptional input as well as mRNAs whose decay rates are affected in OA chondrocytes.
© 2014 The Authors. Arthritis & Rheumatology is published by Wiley Periodicals, Inc. on behalf of the American College of Rheumatology.
Figures
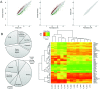
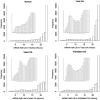


Comment in
-
Editorial: changes in messenger RNA stability in chondrocytes: there's more to osteoarthritis than transcription.Arthritis Rheumatol. 2014 Nov;66(11):2921-3. doi: 10.1002/art.38850. Arthritis Rheumatol. 2014. PMID: 25156089 No abstract available.
-
Osteoarthritis: mRNA decay in OA.Nat Rev Rheumatol. 2014 Oct;10(10):574. doi: 10.1038/nrrheum.2014.155. Epub 2014 Sep 9. Nat Rev Rheumatol. 2014. PMID: 25201381 No abstract available.
References
-
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. - PubMed
-
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, et al. A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–54. - PubMed
-
- Hargrove JL, Schmidt FH. The role of mRNA and protein stability in gene expression. FASEB J. 1989;3:2360–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

