Genome-wide transcriptome analysis shows extensive alternative RNA splicing in the zoonotic parasite Schistosoma japonicum
- PMID: 25156522
- PMCID: PMC4203478
- DOI: 10.1186/1471-2164-15-715
Genome-wide transcriptome analysis shows extensive alternative RNA splicing in the zoonotic parasite Schistosoma japonicum
Abstract
Background: Schistosoma japonicum is a pathogen of the phylum Platyhelminthes that causes zoonotic schistosomiasis in China and Southeast Asian countries where a lack of efficient measures has hampered disease control. The development of tools for diagnosis of acute and chronic infection and for novel antiparasite reagents relies on understanding the biological mechanisms that the parasite exploits.
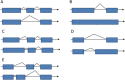
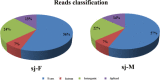
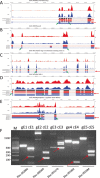
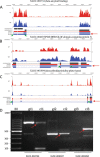
Results: In this study, the polyadenylated transcripts from the male and female S. japonicum were sequenced using a high-throughput RNA-seq technique. Bioinformatic and experimental analyses focused on post-transcriptional RNA processing, which revealed extensive alternative splicing events in the adult stage of the parasite. The numbers of protein-coding sequences identified in the transcriptomes of the female and male S. japonicum were 15,939 and 19,501 respectively, which is more than predicted from the annotated genome sequence. Further, we identified four types of post-transcriptional processing, or alternative splicing, in both female and male worms of S. japonicum: exon skipping, intron retention, and alternative donor and acceptor sites. Unlike mammalian organisms, in S. japonicum, the alternative donor and acceptor sites were more common than the other two types of post-transcriptional processing. In total, respectively 13,438 and 16,507 alternative splicing events were predicted in the transcriptomes of female and male S. japonicum.
Conclusions: By using RNA-seq technology, we obtained the global transcriptomes of male and female S. japonicum. These results further provide a comprehensive view of the global transcriptome of S. japonicum. The findings of a substantial level of alternative splicing events dynamically occurring in S. japonicum parasitization of mammalian hosts suggest complicated transcriptional and post-transcriptional regulation mechanisms employed by the parasite. These data should not only significantly improve the re-annotation of the genome sequences but also should provide new information about the biology of the parasite.
Figures

Similar articles
-
Transcriptome Bioinformatical Analysis of Vertebrate Stages of Schistosoma japonicum Reveals Alternative Splicing Events.PLoS One. 2015 Sep 25;10(9):e0138470. doi: 10.1371/journal.pone.0138470. eCollection 2015. PLoS One. 2015. PMID: 26407301 Free PMC article.
-
Global expression analysis revealed novel gender-specific gene expression features in the blood fluke parasite Schistosoma japonicum.PLoS One. 2011 Apr 6;6(4):e18267. doi: 10.1371/journal.pone.0018267. PLoS One. 2011. PMID: 21494327 Free PMC article.
-
Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum.BMC Genomics. 2010 Jan 21;11:55. doi: 10.1186/1471-2164-11-55. BMC Genomics. 2010. PMID: 20092619 Free PMC article.
-
Integrating transcriptome, proteome and glycome analyses of Schistosoma biology.Trends Parasitol. 2007 Apr;23(4):165-74. doi: 10.1016/j.pt.2007.02.007. Epub 2007 Mar 1. Trends Parasitol. 2007. PMID: 17336161 Review.
-
Schistosoma japonicum infection in the pig as a model for human schistosomiasis japonica.Acta Trop. 2000 Sep 18;76(2):85-99. doi: 10.1016/s0001-706x(00)00103-0. Acta Trop. 2000. PMID: 10936567 Review.
Cited by
-
Genome-wide transcriptome analysis of the early developmental stages of Echinococcus granulosus protoscoleces reveals extensive alternative splicing events in the spliceosome pathway.Parasit Vectors. 2021 Nov 12;14(1):574. doi: 10.1186/s13071-021-05067-9. Parasit Vectors. 2021. PMID: 34772444 Free PMC article.
-
Genome-Wide Analysis of Alternative Splicing Provides Insights into Stress Adaptation of the Pacific Oyster.Mar Biotechnol (NY). 2016 Oct;18(5):598-609. doi: 10.1007/s10126-016-9720-x. Epub 2016 Oct 22. Mar Biotechnol (NY). 2016. PMID: 27771778
-
Non-immune immunoglobulins shield Schistosoma japonicum from host immunorecognition.Sci Rep. 2015 Aug 24;5:13434. doi: 10.1038/srep13434. Sci Rep. 2015. PMID: 26299686 Free PMC article.
-
Comprehensive Transcriptome Analysis of Sex-Biased Expressed Genes Reveals Discrete Biological and Physiological Features of Male and Female Schistosoma japonicum.PLoS Negl Trop Dis. 2016 Apr 29;10(4):e0004684. doi: 10.1371/journal.pntd.0004684. eCollection 2016 Apr. PLoS Negl Trop Dis. 2016. PMID: 27128440 Free PMC article.
-
The Schistosoma mansoni genome encodes thousands of long non-coding RNAs predicted to be functional at different parasite life-cycle stages.Sci Rep. 2017 Sep 5;7(1):10508. doi: 10.1038/s41598-017-10853-6. Sci Rep. 2017. PMID: 28874839 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

