A phosphohistidine proteomics strategy based on elucidation of a unique gas-phase phosphopeptide fragmentation mechanism
- PMID: 25156620
- PMCID: PMC4183637
- DOI: 10.1021/ja507614f
A phosphohistidine proteomics strategy based on elucidation of a unique gas-phase phosphopeptide fragmentation mechanism
Abstract
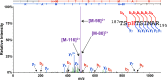
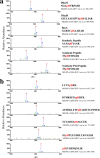
Protein histidine phosphorylation is increasingly recognized as a critical posttranslational modification (PTM) in central metabolism and cell signaling. Still, the detection of phosphohistidine (pHis) in the proteome has remained difficult due to the scarcity of tools to enrich and identify this labile PTM. To address this, we report the first global proteomic analysis of pHis proteins, combining selective immunoenrichment of pHis peptides and a bioinformatic strategy based on mechanistic insight into pHis peptide gas-phase fragmentation during LC-MS/MS. We show that collision-induced dissociation (CID) of pHis peptides produces prominent characteristic neutral losses of 98, 80, and 116 Da. Using isotopic labeling studies, we also demonstrate that the 98 Da neutral loss occurs via gas-phase phosphoryl transfer from pHis to the peptide C-terminal α-carboxylate or to Glu/Asp side chain residues if present. To exploit this property, we developed a software tool that screens LC-MS/MS spectra for potential matches to pHis-containing peptides based on their neutral loss pattern. This tool was integrated into a proteomics workflow for the identification of endogenous pHis-containing proteins in cellular lysates. As an illustration of this strategy, we analyzed pHis peptides from glycerol-fed and mannitol-fed Escherichia coli cells. We identified known and a number of previously speculative pHis sites inferred by homology, predominantly in the phosphoenolpyruvate:sugar transferase system (PTS). Furthermore, we identified two new sites of histidine phosphorylation on aldehyde-alcohol dehydrogenase (AdhE) and pyruvate kinase (PykF) enzymes, previously not known to bear this modification. This study lays the groundwork for future pHis proteomics studies in bacteria and other organisms.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

