Microbial communities evolve faster in extreme environments
- PMID: 25158668
- PMCID: PMC4145313
- DOI: 10.1038/srep06205
Microbial communities evolve faster in extreme environments
Abstract
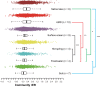
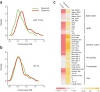
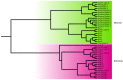
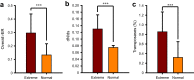
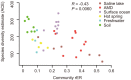
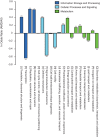
Evolutionary analysis of microbes at the community level represents a new research avenue linking ecological patterns to evolutionary processes, but remains insufficiently studied. Here we report a relative evolutionary rates (rERs) analysis of microbial communities from six diverse natural environments based on 40 metagenomic samples. We show that the rERs of microbial communities are mainly shaped by environmental conditions, and the microbes inhabiting extreme habitats (acid mine drainage, saline lake and hot spring) evolve faster than those populating benign environments (surface ocean, fresh water and soil). These findings were supported by the observation of more relaxed purifying selection and potentially frequent horizontal gene transfers in communities from extreme habitats. The mechanism of high rERs was proposed as high mutation rates imposed by stressful conditions during the evolutionary processes. This study brings us one stage closer to an understanding of the evolutionary mechanisms underlying the adaptation of microbes to extreme environments.
Figures

References
-
- Allen E. E. & Banfield J. F. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 3, 489–498 (2005). - PubMed
-
- Denef V. J., Mueller R. S. & Banfield J. F. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 4, 599–610 (2010). - PubMed
-
- Rothschild L. J. & Mancinelli R. L. Life in extreme environments. Nature 409, 1092–1101 (2001). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

