Keeping it local: evidence for positive selection in Swedish Arabidopsis thaliana
- PMID: 25158800
- PMCID: PMC4209139
- DOI: 10.1093/molbev/msu247
Keeping it local: evidence for positive selection in Swedish Arabidopsis thaliana
Abstract
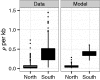
Detecting positive selection in species with heterogeneous habitats and complex demography is notoriously difficult and prone to statistical biases. The model plant Arabidopsis thaliana exemplifies this problem: In spite of the large amounts of data, little evidence for classic selective sweeps has been found. Moreover, many aspects of the demography are unclear, which makes it hard to judge whether the few signals are indeed signs of selection, or false positives caused by demographic events. Here, we focus on Swedish A. thaliana and we find that the demography can be approximated as a two-population model. Careful analysis of the data shows that such a two island model is characterized by a very old split time that significantly predates the last glacial maximum followed by secondary contact with strong migration. We evaluate selection based on this demography and find that this secondary contact model strongly affects the power to detect sweeps. Moreover, it affects the power differently for northern Sweden (more false positives) as compared with southern Sweden (more false negatives). However, even when the demographic history is accounted for, sweep signals in northern Sweden are stronger than in southern Sweden, with little or no positional overlap. Further simulations including the complex demography and selection confirm that this is not compatible with global selection acting on both populations, and thus can be taken as evidence for local selection within subpopulations of Swedish A. thaliana. This study demonstrates the necessity of combining demographic analyses and sweep scans for the detection of selection, particularly when selection acts predominantly local.
Keywords: Arabidopsis thaliana; demography; local adaptation; selective sweeps.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Ågren J, Schemske DW. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 2012;194(4):1112–1122. - PubMed
-
- Beerli P. Effect of unsampled populations on the estimation of population sizes and migration rates between sampled populations. Mol Ecol. 2004;13(4):827–836. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

