In search of the determinants of enhancer-promoter interaction specificity
- PMID: 25160912
- PMCID: PMC4252644
- DOI: 10.1016/j.tcb.2014.07.004
In search of the determinants of enhancer-promoter interaction specificity
Abstract
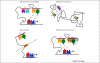
Although it was originally believed that enhancers activate only the nearest promoter, recent global analyses enabled by high-throughput technology suggest that the network of enhancer-promoter interactions is far more complex. The mechanisms that determine the specificity of enhancer-promoter interactions are still poorly understood, but they are thought to include biochemical compatibility, constraints imposed by the three-dimensional architecture of chromosomes, insulator elements, and possibly the effects of local chromatin composition. In this review, we assess the current insights into these determinants, and highlight the functional genomic approaches that will lead the way towards better mechanistic understanding.
Keywords: biochemical compatibility; chromosome architecture; enhancer–promoter interaction; gene regulation.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
References
-
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014;21:210–219. - PubMed
-
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. - PubMed
-
- Lenhard B, et al. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

