PconsFold: improved contact predictions improve protein models
- PMID: 25161237
- PMCID: PMC4147911
- DOI: 10.1093/bioinformatics/btu458
PconsFold: improved contact predictions improve protein models
Abstract
Motivation: Recently it has been shown that the quality of protein contact prediction from evolutionary information can be improved significantly if direct and indirect information is separated. Given sufficiently large protein families, the contact predictions contain sufficient information to predict the structure of many protein families. However, since the first studies contact prediction methods have improved. Here, we ask how much the final models are improved if improved contact predictions are used.
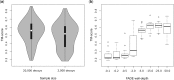
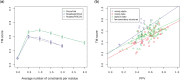
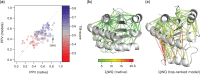
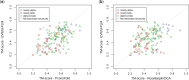
Results: In a small benchmark of 15 proteins, we show that the TM-scores of top-ranked models are improved by on average 33% using PconsFold compared with the original version of EVfold. In a larger benchmark, we find that the quality is improved with 15-30% when using PconsC in comparison with earlier contact prediction methods. Further, using Rosetta instead of CNS does not significantly improve global model accuracy, but the chemistry of models generated with Rosetta is improved.
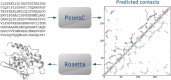
Availability: PconsFold is a fully automated pipeline for ab initio protein structure prediction based on evolutionary information. PconsFold is based on PconsC contact prediction and uses the Rosetta folding protocol. Due to its modularity, the contact prediction tool can be easily exchanged. The source code of PconsFold is available on GitHub at https://www.github.com/ElofssonLab/pcons-fold under the MIT license. PconsC is available from http://c.pcons.net/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

