Ultra-High Density SNParray in Neuroblastoma Molecular Diagnostics
- PMID: 25161957
- PMCID: PMC4129917
- DOI: 10.3389/fonc.2014.00202
Ultra-High Density SNParray in Neuroblastoma Molecular Diagnostics
Abstract
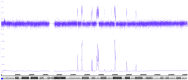
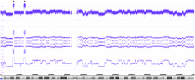
Neuroblastoma serves as a paradigm for applying tumor genomic data for determining patient prognosis and thus for treatment allocation. MYCN status, i.e., amplified vs. non-amplified, was one of the very first biomarkers in oncology to discriminate aggressive from less aggressive or even favorable clinical courses of neuroblastoma. However, MYCN amplification is by far not the only genetic change associated with unfavorable clinical courses. So called "segmental chromosomal aberrations," (SCAs) i.e., gains or losses of chromosomal fragments, can also indicate tumor aggressiveness. The clinical use of these genomic aberrations has, however, been hampered for many years by methodical and interpretational problems. Only after reaching worldwide consensus on markers, methodology, and data interpretation, information on SCAs has recently been implemented in clinical studies. Now, a number of collaborative studies within COG, GPOH, and SIOPEN use genomic information to stratify therapy for patients with localized and metastatic disease. Recently, new types of DNA based aberrations influencing the clinical behavior of neuroblastomas have been described. Deletions or mutations of genes like ATRX and a phenomenon referred to as "chromothripsis" are all assumed to correlate with an unfavorable clinical behavior. However, these genomic aberrations need to be scrutinized in larger studies applying the most appropriate techniques. Single nucleotide polymorphism arrays have proven successful in deciphering genomic aberrations of cancer cells; these techniques, however, are usually not applied in the daily routine. Here, we present an ultra-high density (UHD) SNParray technique which is, because of its high specificity and sensitivity and the combined copy number and allele information, highly appropriate for the genomic diagnosis of neuroblastoma and other malignancies.
Keywords: INRG; SNParray; amplification; chromothripsis; genetic risk factors; genomic; neuroblastoma; precision medicine.
Figures
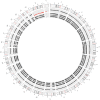
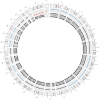
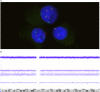
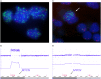


References
-
- Evans AE, Gerson J, Schnaufer L. Spontaneous regression of neuroblastoma. Natl Cancer Inst Monogr (1976) 44:49–54 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

