Investigating the host specificity of Campylobacter jejuni and Campylobacter coli by sequencing gyrase subunit A
- PMID: 25163418
- PMCID: PMC4156964
- DOI: 10.1186/s12866-014-0205-7
Investigating the host specificity of Campylobacter jejuni and Campylobacter coli by sequencing gyrase subunit A
Abstract
Background: Surveillance and field investigations of Campylobacter infections require molecular tools with genetic markers appropriate for tracing purposes, i.e. based on the principle that some Campylobacter lineages acquire a host signature under adaptive selection pressure. We developed a sequence-based method targeting the quinolone resistance determining region within the subunit A of DNA gyrase (gyrA). Host specificity was evaluated by characterizing two collections of Campylobacter jejuni (N = 430) and Campylobacter coli (N = 302) originating from surface waters, domestic mammals and poultry.
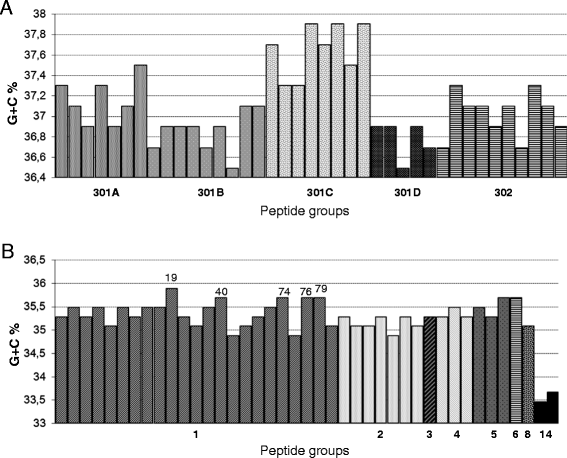
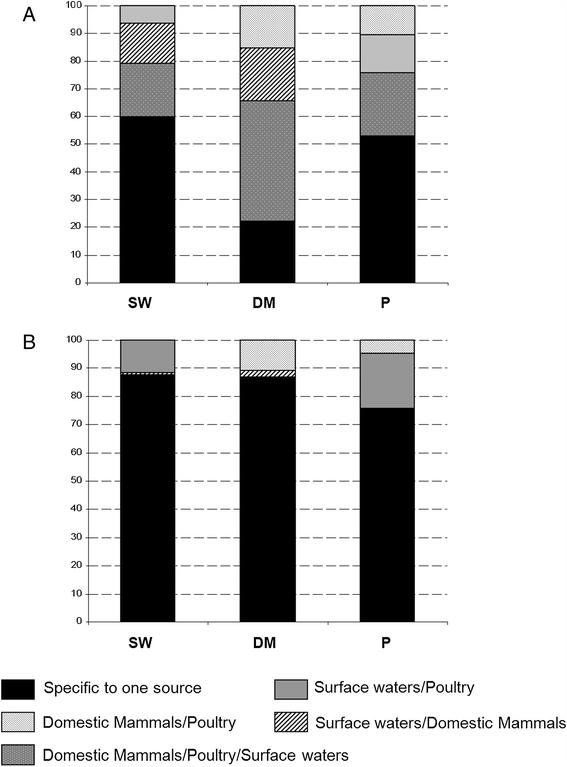
Results: Based on nucleotide identity, a total of 80 gyrA alleles were observed. Thirty nine alleles assigned to C. coli encoding two peptides fell into three clades: two associated with surface waters and one associated with domestic mammals and poultry. The variability in GC content generated by synonymous mutations suggested that surface waters isolates originated from two distinct ecological niches. A total of 42 alleles were recorded from C. jejuni strains and encoded 8 peptides including one lying in a distinct lineage associated with wildlife. Seven of the 23 alleles encoding peptide #1 displayed the synonymous mutation G408A not identified in poultry isolates. By contrast, the substitution Ser22Gly observed in 4 different peptide groups was significantly associated with domestic birds (P = 0.001). The change in amino acid sequences Thr86Ile conferring resistance to quinolones was significantly associated with poultry (P < 0.001) in both C. jejuni and C. coli with 38.7% and 67.9% of quinolone-resistant strains, respectively.
Conclusions: The gyrA typing method presented here is an informative tool as sequences appear to be predictive of particular ecological niches. Combined with multi-locus sequence typing, it could increase the resolution of source attribution, and combined with porA/flaA typing it could be suitable for detecting temporal clusters of human cases. All gyrA alleles identified were deposited in the freely accessible online database http://pubmlst.org/campylobacter.
Figures

References
-
- EFSA, (European Food Safety Authority), ECDC, (European Centre for Disease Prevention and Control) The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2011. EFSA J. 2013;11(4):3129.
-
- Sécurité alimentaire/Luxembourg - Rapports d’Activité - OSQCA.; [http://www.securite-alimentaire.public.lu/organisme/rapports_activite_os...]
-
- Ragimbeau C, Schneider F, Losch S, Even J, Mossong J. Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl Environ Microbiol. 2008;74:7715–7722. doi: 10.1128/AEM.00865-08. - DOI - PMC - PubMed
-
- Mughini Gras L, Smid JH, Wagenaar JA, De Boer AG, Havelaar AH, Friesema IHM, French NP, Busani L, Van Pelt W. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: a combined case–control and source attribution analysis. PLoS One. 2012;7:e42599. doi: 10.1371/journal.pone.0042599. - DOI - PMC - PubMed
-
- Strachan NJC, Gormley FJ, Rotariu O, Ogden ID, Miller G, Dunn GM, Sheppard SK, Dallas JF, Reid TMS, Howie H, Maiden MCJ, Forbes KJ. Attribution of campylobacter infections in Northeast Scotland to specific sources by use of multilocus sequence typing. J Infect Dis. 2009;199:1205–1208. doi: 10.1086/597417. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

