Genome-driven integrated classification of breast cancer validated in over 7,500 samples
- PMID: 25164602
- PMCID: PMC4166472
- DOI: 10.1186/s13059-014-0431-1
Genome-driven integrated classification of breast cancer validated in over 7,500 samples
Abstract
Background: IntClust is a classification of breast cancer comprising 10 subtypes based on molecular drivers identified through the integration of genomic and transcriptomic data from 1,000 breast tumors and validated in a further 1,000. We present a reliable method for subtyping breast tumors into the IntClust subtypes based on gene expression and demonstrate the clinical and biological validity of the IntClust classification.
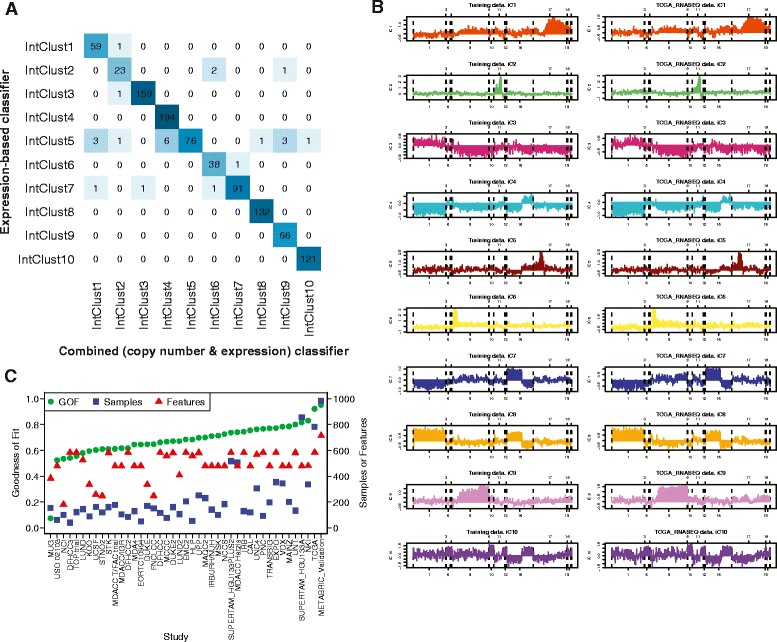
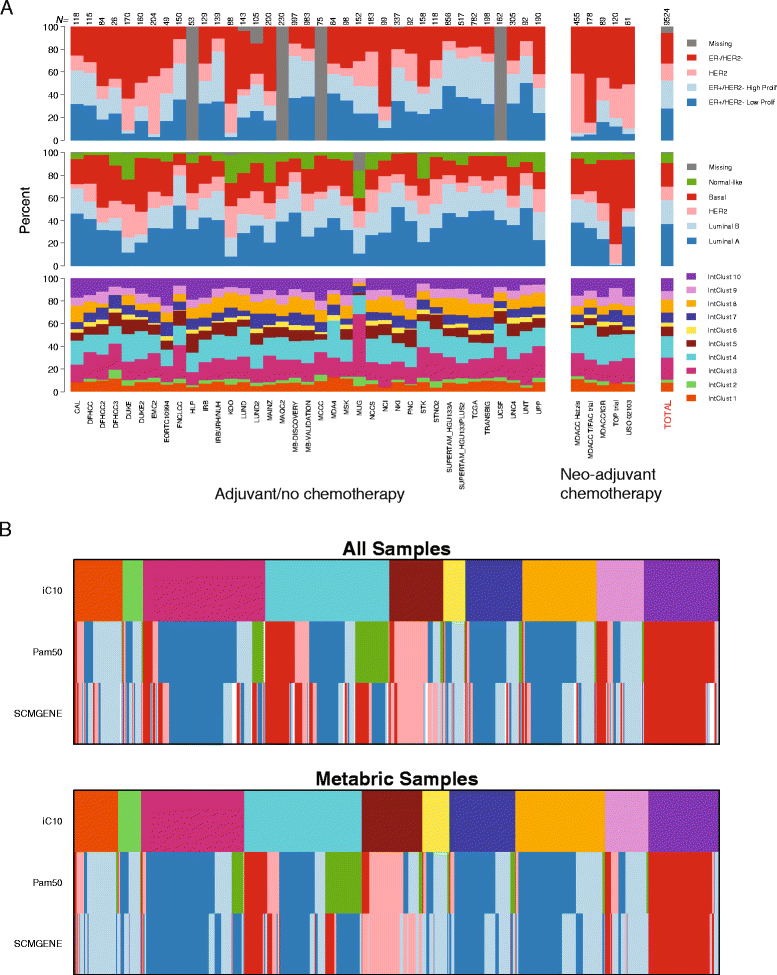
Results: We developed a gene expression-based approach for classifying breast tumors into the ten IntClust subtypes by using the ensemble profile of the index discovery dataset. We evaluate this approach in 983 independent samples for which the combined copy-number and gene expression IntClust classification was available. Only 24 samples are discordantly classified. Next, we compile a consolidated external dataset composed of a further 7,544 breast tumors. We use our approach to classify all samples into the IntClust subtypes. All ten subtypes are observable in most studies at comparable frequencies. The IntClust subtypes are significantly associated with relapse-free survival and recapitulate patterns of survival observed previously. In studies of neo-adjuvant chemotherapy, IntClust reveals distinct patterns of chemosensitivity. Finally, patterns of expression of genomic drivers reported by TCGA (The Cancer Genome Atlas) are better explained by IntClust as compared to the PAM50 classifier.
Conclusions: IntClust subtypes are reproducible in a large meta-analysis, show clinical validity and best capture variation in genomic drivers. IntClust is a driver-based breast cancer classification and is likely to become increasingly relevant as more targeted biological therapies become available.
Figures


References
-
- Perou C, Sørlie T, Eisen M, van de Rijn M, Jeffrey S, Rees C, Pollack J, Ross D, Johnsen H, Akslen L, Fluge O, Pergamenschikov A, Williams C, Zhu S, Lønning P, Børresen-Dale A, Brown P, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI - PubMed
-
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. - DOI - PMC - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. - DOI - PubMed
-
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

