Comparative analysis of the transcriptome across distant species
- PMID: 25164755
- PMCID: PMC4155737
- DOI: 10.1038/nature13424
Comparative analysis of the transcriptome across distant species
Abstract
The transcriptome is the readout of the genome. Identifying common features in it across distant species can reveal fundamental principles. To this end, the ENCODE and modENCODE consortia have generated large amounts of matched RNA-sequencing data for human, worm and fly. Uniform processing and comprehensive annotation of these data allow comparison across metazoan phyla, extending beyond earlier within-phylum transcriptome comparisons and revealing ancient, conserved features. Specifically, we discover co-expression modules shared across animals, many of which are enriched in developmental genes. Moreover, we use expression patterns to align the stages in worm and fly development and find a novel pairing between worm embryo and fly pupae, in addition to the embryo-to-embryo and larvae-to-larvae pairings. Furthermore, we find that the extent of non-canonical, non-coding transcription is similar in each organism, per base pair. Finally, we find in all three organisms that the gene-expression levels, both coding and non-coding, can be quantitatively predicted from chromatin features at the promoter using a 'universal model' based on a single set of organism-independent parameters.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



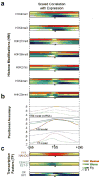
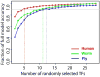


Comment in
-
Genomics: Hiding in plain sight.Nature. 2014 Aug 28;512(7515):374-5. doi: 10.1038/512374a. Nature. 2014. PMID: 25164742 No abstract available.
References
-
- Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–8. - PubMed
-
- Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–93. - PubMed
-
- Levin M, Hashimshony T, Wagner F, Yanai I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell. 2012;22:1101–8. - PubMed
-
- Kalinka AT, et al. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HG004271/HG/NHGRI NIH HHS/United States
- R01 GM076655/GM/NIGMS NIH HHS/United States
- T15 LM007056/LM/NLM NIH HHS/United States
- U54 HG007004/HG/NHGRI NIH HHS/United States
- HG007000/HG/NHGRI NIH HHS/United States
- U41 HG007355/HG/NHGRI NIH HHS/United States
- 5U01HG004695-04/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54 HG007005/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U01 HG007031/HG/NHGRI NIH HHS/United States
- T32 HD060555/HD/NICHD NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- U01 HG004261/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U54 HG006994/HG/NHGRI NIH HHS/United States
- K99 HG006698/HG/NHGRI NIH HHS/United States
- 5U54HG004555/HG/NHGRI NIH HHS/United States
- U01 HG 004263/HG/NHGRI NIH HHS/United States
- U01-HG004261/HG/NHGRI NIH HHS/United States
- RC2-HG005639/HG/NHGRI NIH HHS/United States
- U54HG007005/HG/NHGRI NIH HHS/United States
- HG007355/HG/NHGRI NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- U54 HG006944/HG/NHGRI NIH HHS/United States
- U01HG004258/HG/NHGRI NIH HHS/United States
- U01 HG004258/HG/NHGRI NIH HHS/United States
- 1U01HG007031-01/HG/NHGRI NIH HHS/United States
- U01 HG004695/HG/NHGRI NIH HHS/United States
- U41 HG007000/HG/NHGRI NIH HHS/United States
- U54 HG004555/HG/NHGRI NIH HHS/United States
- WT098051/WT_/Wellcome Trust/United Kingdom
- RC2 HG005639/HG/NHGRI NIH HHS/United States
- ZIA DK015600/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

