LoRDEC: accurate and efficient long read error correction
- PMID: 25165095
- PMCID: PMC4253826
- DOI: 10.1093/bioinformatics/btu538
LoRDEC: accurate and efficient long read error correction
Abstract
Motivation: PacBio single molecule real-time sequencing is a third-generation sequencing technique producing long reads, with comparatively lower throughput and higher error rate. Errors include numerous indels and complicate downstream analysis like mapping or de novo assembly. A hybrid strategy that takes advantage of the high accuracy of second-generation short reads has been proposed for correcting long reads. Mapping of short reads on long reads provides sufficient coverage to eliminate up to 99% of errors, however, at the expense of prohibitive running times and considerable amounts of disk and memory space.
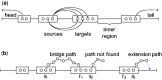
Results: We present LoRDEC, a hybrid error correction method that builds a succinct de Bruijn graph representing the short reads, and seeks a corrective sequence for each erroneous region in the long reads by traversing chosen paths in the graph. In comparison, LoRDEC is at least six times faster and requires at least 93% less memory or disk space than available tools, while achieving comparable accuracy. Availability and implementaion: LoRDEC is written in C++, tested on Linux platforms and freely available at http://atgc.lirmm.fr/lordec.
© The Author 2014. Published by Oxford University Press.
Figures



Similar articles
-
Evaluation and Validation of Assembling Corrected PacBio Long Reads for Microbial Genome Completion via Hybrid Approaches.PLoS One. 2015 Dec 7;10(12):e0144305. doi: 10.1371/journal.pone.0144305. eCollection 2015. PLoS One. 2015. PMID: 26641475 Free PMC article.
-
A hybrid and scalable error correction algorithm for indel and substitution errors of long reads.BMC Genomics. 2019 Dec 20;20(Suppl 11):948. doi: 10.1186/s12864-019-6286-9. BMC Genomics. 2019. PMID: 31856721 Free PMC article.
-
Accurate self-correction of errors in long reads using de Bruijn graphs.Bioinformatics. 2017 Mar 15;33(6):799-806. doi: 10.1093/bioinformatics/btw321. Bioinformatics. 2017. PMID: 27273673 Free PMC article.
-
A comprehensive evaluation of long read error correction methods.BMC Genomics. 2020 Dec 21;21(Suppl 6):889. doi: 10.1186/s12864-020-07227-0. BMC Genomics. 2020. PMID: 33349243 Free PMC article. Review.
-
The present and future of de novo whole-genome assembly.Brief Bioinform. 2018 Jan 1;19(1):23-40. doi: 10.1093/bib/bbw096. Brief Bioinform. 2018. PMID: 27742661 Review.
Cited by
-
SPAligner: alignment of long diverged molecular sequences to assembly graphs.BMC Bioinformatics. 2020 Jul 24;21(Suppl 12):306. doi: 10.1186/s12859-020-03590-7. BMC Bioinformatics. 2020. PMID: 32703258 Free PMC article.
-
Characterization and Analysis of the Full-Length Transcriptomes of Multiple Organs in Pseudotaxus chienii (W.C.Cheng) W.C.Cheng.Int J Mol Sci. 2020 Jun 17;21(12):4305. doi: 10.3390/ijms21124305. Int J Mol Sci. 2020. PMID: 32560294 Free PMC article.
-
Draft genomic sequence of Armillaria gallica 012m: insights into its symbiotic relationship with Gastrodia elata.Braz J Microbiol. 2020 Dec;51(4):1539-1552. doi: 10.1007/s42770-020-00317-x. Epub 2020 Jun 22. Braz J Microbiol. 2020. PMID: 32572836 Free PMC article.
-
Sequence data for Clostridium autoethanogenum using three generations of sequencing technologies.Sci Data. 2015 Apr 14;2:150014. doi: 10.1038/sdata.2015.14. eCollection 2015. Sci Data. 2015. PMID: 25977818 Free PMC article.
-
Comprehensive profiling of epigenetic modifications in fast-growing Moso bamboo shoots.Plant Physiol. 2023 Feb 12;191(2):1017-1035. doi: 10.1093/plphys/kiac525. Plant Physiol. 2023. PMID: 36417282 Free PMC article.
References
-
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Cazaux B, et al. CPM, volume 8486 of LNCS. Springer; 2014. From indexing data structures to de bruijn graphs; pp. 89–99.
-
- Chaisson M, et al. Fragment assembly with short reads. Bioinformatics. 2004;20:2067–2074. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

