Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex
- PMID: 25165108
- PMCID: PMC4248913
- DOI: 10.1128/JVI.02139-14
Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex
Abstract
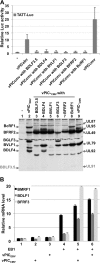
During their productive cycle, herpesviruses exhibit a strictly regulated temporal cascade of gene expression that has three general stages: immediate early (IE), early (E), and late (L). Promoter complexity differs strikingly between IE/E genes and L genes. IE and E promoters contain cis-regulating sequences upstream of a TATA box, whereas L promoters comprise a unique cis element. In the case of the gammaherpesviruses, this element is usually a TATT motif found in the position where the consensus TATA box of eukaryotic promoters is typically found. Epstein-Barr virus (EBV) encodes a protein, called BcRF1, which has structural homology with the TATA-binding protein and interacts specifically with the TATT box. However, although necessary for the expression of the L genes, BcRF1 is not sufficient, suggesting that other viral proteins are also required. Here, we present the identification and characterization of a viral protein complex necessary and sufficient for the expression of the late viral genes. This viral complex is composed of five different proteins in addition to BcRF1 and interacts with cellular RNA polymerase II. During the viral productive cycle, this complex, which we call the vPIC (for viral preinitiation complex), works in concert with the viral DNA replication machinery to activate expression of the late viral genes. The EBV vPIC components have homologs in beta- and gammaherpesviruses but not in alphaherpesviruses. Our results not only reveal that beta- and gammaherpesviruses encode their own transcription preinitiation complex responsible for the expression of the late viral genes but also indicate the close evolutionary history of these viruses.
Importance: Control of late gene transcription in DNA viruses is a major unsolved question in virology. In eukaryotes, the first step in transcriptional activation is the formation of a permissive chromatin, which allows assembly of the preinitiation complex (PIC) at the core promoter. Fixation of the TATA box-binding protein (TBP) is a key rate-limiting step in this process. This study provides evidence that EBV encodes a complex composed of six proteins necessary for the expression of the late viral genes. This complex is formed around a viral TBP-like protein and interacts with cellular RNA polymerase II, suggesting that it is directly involved in the assembly of a virus-specific PIC (vPIC).
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Rickinson AB, Kieff E. 2001. Epstein-Barr virus, p 2575–2627 In Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE. (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

