HIV control is mediated in part by CD8+ T-cell targeting of specific epitopes
- PMID: 25165115
- PMCID: PMC4249072
- DOI: 10.1128/JVI.01004-14
HIV control is mediated in part by CD8+ T-cell targeting of specific epitopes
Abstract
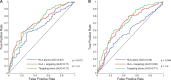
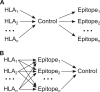
We investigated the hypothesis that the correlation between the class I HLA types of an individual and whether that individual spontaneously controls HIV-1 is mediated by the targeting of specific epitopes by CD8(+) T cells. By measuring gamma interferon enzyme-linked immunosorbent spot (ELISPOT) assay responses to a panel of 257 optimally defined epitopes in 341 untreated HIV-infected persons, including persons who spontaneously control viremia, we found that the correlation between HLA types and control is mediated by the targeting of specific epitopes. Moreover, we performed a graphical model-based analysis that suggested that the targeting of specific epitopes is a cause of such control--that is, some epitopes are protective rather than merely associated with control--and identified eight epitopes that are significantly protective. In addition, we use an in silico analysis to identify protein regions where mutations are likely to affect the stability of a protein, and we found that the protective epitopes identified by the ELISPOT analysis correspond almost perfectly to such regions. This in silico analysis thus suggests a possible mechanism for control and could be used to identify protective epitopes that are not often targeted in natural infection but that may be potentially useful in a vaccine. Our analyses thus argue for the inclusion (and exclusion) of specific epitopes in an HIV vaccine.
Importance: Some individuals naturally control HIV replication in the absence of antiretroviral therapy, and this ability to control is strongly correlated with the HLA class I alleles that they express. Here, in a large-scale experimental study, we provide evidence that this correlation is mediated largely by the targeting of specific CD8(+) T-cell epitopes, and we identify eight epitopes that are likely to cause control. In addition, we provide an in silico analysis indicating that control occurs because mutations within these epitopes change the stability of the protein structures. This in silico analysis also identified additional epitopes that are not typically targeted in natural infection but may lead to control when included in a vaccine, provided that other epitopes that would otherwise distract the immune system from targeting them are excluded from the vaccine.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Kaslow RA, Carrington M, Apple R, Park L, Muñoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411. 10.1038/nm0496-405. - DOI - PubMed
-
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal SA, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947. 10.1126/science.1143767. - DOI - PMC - PubMed
-
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PIW, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DML, Vine S, Addo MM, Allen TM, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. 10.1126/science.1195271. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

