The novel component Kgd4 recruits the E3 subunit to the mitochondrial α-ketoglutarate dehydrogenase
- PMID: 25165143
- PMCID: PMC4214781
- DOI: 10.1091/mbc.E14-07-1178
The novel component Kgd4 recruits the E3 subunit to the mitochondrial α-ketoglutarate dehydrogenase
Abstract
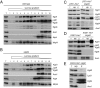
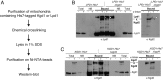
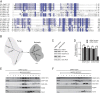
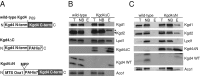
The mitochondrial citric acid cycle is a central hub of cellular metabolism, providing intermediates for biosynthetic pathways and channeling electrons to the respiratory chain complexes. In this study, we elucidated the composition and organization of the multienzyme complex α-ketoglutarate dehydrogenase (α-KGDH). In addition to the three classical E1-E3 subunits, we identified a novel component, Kgd4 (Ymr31/MRPS36), which was previously assigned to be a subunit of the mitochondrial ribosome. Biochemical analyses demonstrate that this protein plays an evolutionarily conserved role in the organization of mitochondrial α-KGDH complexes of fungi and animals. By binding to both the E1-E2 core and the E3 subunit, Kgd4 acts as a molecular adaptor that is necessary to a form a stable α-KGDH enzyme complex. Our work thus reveals a novel subunit of a key citric acid-cycle enzyme and shows how this large complex is organized.
© 2014 Heublein et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures




References
-
- Aevarsson A, Seger K, Turley S, Sokatch JR, Hol WG. Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes. Nat Struct Biol. 1999;6:785–792. - PubMed
-
- Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

