Classical mathematical models for description and prediction of experimental tumor growth
- PMID: 25167199
- PMCID: PMC4148196
- DOI: 10.1371/journal.pcbi.1003800
Classical mathematical models for description and prediction of experimental tumor growth
Abstract
Despite internal complexity, tumor growth kinetics follow relatively simple laws that can be expressed as mathematical models. To explore this further, quantitative analysis of the most classical of these were performed. The models were assessed against data from two in vivo experimental systems: an ectopic syngeneic tumor (Lewis lung carcinoma) and an orthotopically xenografted human breast carcinoma. The goals were threefold: 1) to determine a statistical model for description of the measurement error, 2) to establish the descriptive power of each model, using several goodness-of-fit metrics and a study of parametric identifiability, and 3) to assess the models' ability to forecast future tumor growth. The models included in the study comprised the exponential, exponential-linear, power law, Gompertz, logistic, generalized logistic, von Bertalanffy and a model with dynamic carrying capacity. For the breast data, the dynamics were best captured by the Gompertz and exponential-linear models. The latter also exhibited the highest predictive power, with excellent prediction scores (≥80%) extending out as far as 12 days in the future. For the lung data, the Gompertz and power law models provided the most parsimonious and parametrically identifiable description. However, not one of the models was able to achieve a substantial prediction rate (≥70%) beyond the next day data point. In this context, adjunction of a priori information on the parameter distribution led to considerable improvement. For instance, forecast success rates went from 14.9% to 62.7% when using the power law model to predict the full future tumor growth curves, using just three data points. These results not only have important implications for biological theories of tumor growth and the use of mathematical modeling in preclinical anti-cancer drug investigations, but also may assist in defining how mathematical models could serve as potential prognostic tools in the clinic.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
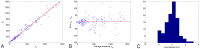
 against approximation of the volume given by the average of the two measurement
against approximation of the volume given by the average of the two measurement  . The χ
2 test rejected Gaussian distribution of constant variance (
. The χ
2 test rejected Gaussian distribution of constant variance ( ) C. Histogram of the normalized error
) C. Histogram of the normalized error  applying the error model given by
applying the error model given by  with α = 0.84 and Vm = 83 mm3. It shows Gaussian distribution (χ
2 test not rejected, p = 0.196) with standard deviation
with α = 0.84 and Vm = 83 mm3. It shows Gaussian distribution (χ
2 test not rejected, p = 0.196) with standard deviation  .
.
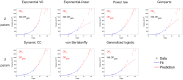

 , as quantified by the score
, as quantified by the score  (multiplied by 100), defined in (17). This proportion only includes animals having measurements at these two time points, thus values at different row d on the same column n or reverse might represent predictions in different animals. White squares correspond to situations where this number was too low (<5) and thus success score, considered not significant, was not reported. Bottom: distribution of the relative error of prediction (20), all animals and (n,d) pooled together. Models were ranked in ascending order of overall mean success score reported in Tables 5 and 6. A. Lung tumor data. B. Breast tumor data.
(multiplied by 100), defined in (17). This proportion only includes animals having measurements at these two time points, thus values at different row d on the same column n or reverse might represent predictions in different animals. White squares correspond to situations where this number was too low (<5) and thus success score, considered not significant, was not reported. Bottom: distribution of the relative error of prediction (20), all animals and (n,d) pooled together. Models were ranked in ascending order of overall mean success score reported in Tables 5 and 6. A. Lung tumor data. B. Breast tumor data.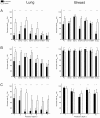
 (see Materials and Methods, Models predictions methods for its definition). B. Benefit of the method for prediction of the next day, using three data points (score
(see Materials and Methods, Models predictions methods for its definition). B. Benefit of the method for prediction of the next day, using three data points (score  ). C. Prediction improvement at various prediction depths, using the power law model (lung data) or the exponential-linear model (breast data). Due to lack of animals to be predicted for some of the random assignments, results of depths 2, 4, 6 and 9 for the breast data were not considered significant and were not reported (see Materials and Methods). * = p<0.05, ** = p<0.001, Student's t-test.
). C. Prediction improvement at various prediction depths, using the power law model (lung data) or the exponential-linear model (breast data). Due to lack of animals to be predicted for some of the random assignments, results of depths 2, 4, 6 and 9 for the breast data were not considered significant and were not reported (see Materials and Methods). * = p<0.05, ** = p<0.001, Student's t-test.References
-
- Talmadge JE, Singh RK, Fidler IJ, Raz A (2007) Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol 170: 793–804 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1864878&tool=p.... Accessed 24 January 2014. - PMC - PubMed
-
- Ebos JML, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, et al. (2009) Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell 15: 232–239 Available: http://www.ncbi.nlm.nih.gov/pubmed/19249681. Accessed 14 July 2012. - PMC - PubMed
-
- Collins VP, Loeffler RK, Tivey H (1956) Observations on growth rates of human tumors. Am J Roentgenol Radium Ther Nucl Med 76: 988–1000. - PubMed
-
- Steel GG (1977) Growth kinetics of tumours. Clarendon Press. Oxford.
-
- Hart D, Shochat E, Agur Z (1998) The growth law of primary breast cancer as inferred from mammography screening trials data. Br J Cancer 78: 382–387 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2063020&tool=p.... - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

