Comprehensive analysis of pathogenic deletion variants in Fanconi anemia genes
- PMID: 25168418
- PMCID: PMC4407816
- DOI: 10.1002/humu.22680
Comprehensive analysis of pathogenic deletion variants in Fanconi anemia genes
Abstract
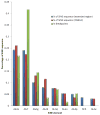
Fanconi anemia (FA) is a rare recessive disease resulting from mutations in one of at least 16 different genes. Mutation types and phenotypic manifestations of FA are highly heterogeneous and influence the clinical management of the disease. We analyzed 202 FA families for large deletions, using high-resolution comparative genome hybridization arrays, single-nucleotide polymorphism arrays, and DNA sequencing. We found pathogenic deletions in 88 FANCA, seven FANCC, two FANCD2, and one FANCB families. We find 35% of FA families carry large deletions, accounting for 18% of all FA pathogenic variants. Cloning and sequencing across the deletion breakpoints revealed that 52 FANCA deletion ends, and one FANCC deletion end extended beyond the gene boundaries, potentially affecting neighboring genes with phenotypic consequences. Seventy-five percent of the FANCA deletions are Alu-Alu mediated, predominantly by AluY elements, and appear to be caused by nonallelic homologous recombination. Individual Alu hotspots were identified. Defining the haplotypes of four FANCA deletions shared by multiple families revealed that three share a common ancestry. Knowing the exact molecular changes that lead to the disease may be critical for a better understanding of the FA phenotype, and to gain insight into the mechanisms driving these pathogenic deletion variants.
Keywords: FANCA; FANCB; FANCC; FANCD2; Fanconi anemia; arrayCGH.
© 2014 WILEY PERIODICALS, INC.
Conflict of interest statement
Figures


References
-
- Ameziane N, Errami A, Leveille F, Fontaine C, de Vries Y, van Spaendonk RM, de Winter JP, Pals G, Joenje H. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum Mutat. 2008;29:159–66. - PubMed
-
- Amouri A, Talmoudi F, Messaoud O, d’Enghien CD, Rekaya MB, Allegui I, Azaiez H, Kefi R, Abdelhak A, Meseddi SH, et al. High frequency of exon 15 deletion in the FANCA gene in Tunisian patients affected with Fanconi anemia disease: implication for diagnosis. Mol Genet Genomic Med. 2014;2:160–5. - PMC - PubMed
-
- Auerbach AD, Greenbaum J, Pujara K, Batish SD, Bitencourt MA, Kokemohr I, Schneider H, Lobitzc S, Pasquini R, Giampietro PF, et al. Spectrum of sequence variation in the FANCG gene: an International Fanconi Anemia Registry (IFAR) study. Hum Mutat. 2003;21:158–68. - PubMed
-
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

