Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family
- PMID: 25169981
- PMCID: PMC4202323
- DOI: 10.1093/gbe/evu187
Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family
Abstract
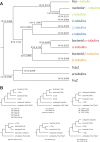
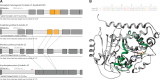
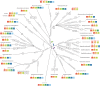
Tubulins belong to the most abundant proteins in eukaryotes providing the backbone for many cellular substructures like the mitotic and meiotic spindles, the intracellular cytoskeletal network, and the axonemes of cilia and flagella. Homologs have even been reported for archaea and bacteria. However, a taxonomically broad and whole-genome-based analysis of the tubulin protein family has never been performed, and thus, the number of subfamilies, their taxonomic distribution, and the exact grouping of the supposed archaeal and bacterial homologs are unknown. Here, we present the analysis of 3,524 tubulins from 504 species. The tubulins formed six major subfamilies, α to ζ. Species of all major kingdoms of the eukaryotes encode members of these subfamilies implying that they must have already been present in the last common eukaryotic ancestor. The proposed archaeal homologs grouped together with the bacterial TubZ proteins as sister clade to the FtsZ proteins indicating that tubulins are unique to eukaryotes. Most species contained α- and/or β-tubulin gene duplicates resulting from recent branch- and species-specific duplication events. This shows that tubulins cannot be used for constructing species phylogenies without resolving their ortholog-paralog relationships. The many gene duplicates and also the independent loss of the δ-, ε-, or ζ-tubulins, which have been shown to be part of the triplet microtubules in basal bodies, suggest that tubulins can functionally substitute each other.
Keywords: FtsZ; TubZ; artubulin; eukaryotic evolution; gene duplication; tubulin.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Aylett CHS, Löwe J, Amos LA. New insights into the mechanisms ofcytomotive actin and tubulin filaments. Int Rev Cell Mol Biol. 2011;292:1–71. - PubMed
-
- Breviario D, Gianì S, Morello L. Multiple tubulins: evolutionary aspects and biological implications. Plant J Cell Mol Biol. 2013;75:202–218. - PubMed
-
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

