VSEAMS: a pipeline for variant set enrichment analysis using summary GWAS data identifies IKZF3, BATF and ESRRA as key transcription factors in type 1 diabetes
- PMID: 25170024
- PMCID: PMC4296156
- DOI: 10.1093/bioinformatics/btu571
VSEAMS: a pipeline for variant set enrichment analysis using summary GWAS data identifies IKZF3, BATF and ESRRA as key transcription factors in type 1 diabetes
Abstract
Motivation: Genome-wide association studies (GWAS) have identified many loci implicated in disease susceptibility. Integration of GWAS summary statistics (P-values) and functional genomic datasets should help to elucidate mechanisms.
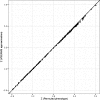
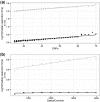
Results: We extended a non-parametric SNP set enrichment method to test for enrichment of GWAS signals in functionally defined loci to a situation where only GWAS P-values are available. The approach is implemented in VSEAMS, a freely available software pipeline. We use VSEAMS to identify enrichment of type 1 diabetes (T1D) GWAS associations near genes that are targets for the transcription factors IKZF3, BATF and ESRRA. IKZF3 lies in a known T1D susceptibility region, while BATF and ESRRA overlap other immune disease susceptibility regions, validating our approach and suggesting novel avenues of research for T1D.
Availability and implementation: VSEAMS is available for download (http://github.com/ollyburren/vseams).
© The Author 2014. Published by Oxford University Press.
Figures



References
-
- Geweke J. Bayesian Statistics 4: Evaluating the Accuracy of Sampling-based Approaches to the Calculation of Posterior Moments. Oxford, UK: Oxford University Press; 1992.
Publication types
MeSH terms
Substances
Grants and funding
- MH059571/MH/NIMH NIH HHS/United States
- MH067257/MH/NIMH NIH HHS/United States
- 076113/WT_/Wellcome Trust/United Kingdom
- R01 MH067257/MH/NIMH NIH HHS/United States
- R01 MH059587/MH/NIMH NIH HHS/United States
- MH059588/MH/NIMH NIH HHS/United States
- MH59566/MH/NIMH NIH HHS/United States
- MH59587/MH/NIMH NIH HHS/United States
- R01 MH061675/MH/NIMH NIH HHS/United States
- MH061675/MH/NIMH NIH HHS/United States
- MH60870/MH/NIMH NIH HHS/United States
- MH59586/MH/NIMH NIH HHS/United States
- R01 MH060870/MH/NIMH NIH HHS/United States
- U01 DK062418/DK/NIDDK NIH HHS/United States
- R01 MH059571/MH/NIMH NIH HHS/United States
- R01 MH059565/MH/NIMH NIH HHS/United States
- MH059565/MH/NIMH NIH HHS/United States
- R01 MH059586/MH/NIMH NIH HHS/United States
- R01 MH059566/MH/NIMH NIH HHS/United States
- 091157/Wellcome Trust/United Kingdom
- R01 MH063420/MH/NIMH NIH HHS/United States
- R01 MH059588/MH/NIMH NIH HHS/United States
- U01 MH060879/MH/NIMH NIH HHS/United States
- R01 MH060879/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

