MicroRNAs in kidney transplantation
- PMID: 25170095
- PMCID: PMC4301831
- DOI: 10.1093/ndt/gfu280
MicroRNAs in kidney transplantation
Abstract
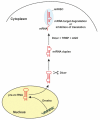
The discovery of novel classes of non-coding RNAs (ncRNAs) has revolutionized medicine. Long thought to be a mere cellular housekeeper, surprising functions have recently been uncovered. MicroRNAs (miRNAs), are a representative of the class of short ncRNAs, play a fundamental role in the control of DNA and protein biosynthesis and activity as well as pathology. Currently, miRNAs are being investigated as diagnostic and prognostic markers and potential therapeutic targets in kidney transplantation for such indolent processes as ischaemia-reperfusion injury, humoral rejection or viral infections. It is realistic to believe that monitoring of renal allograft recipients in the future will include genome-wide miRNA profiling of biological fluids. Based on these individual profiles, an informed decision on therapeutic consequences will be possible. A first success with a specific suppression of miRNAs by antisense oligonucleotides was achieved in experimental studies of reperfusion injury and humoral rejection. Proof of this concept in men comes from studies in such indolent viral infections as Ebola and hepatitis C, where anti-miR therapy led to sustained viral clearance. In this review, we summarize the basis of the recent ncRNA revolution and its implication for kidney transplantation.
Keywords: acute kidney injury; biomarker; miRNAs; rejection; renal transplantation.
© The Author 2014. Published by Oxford University Press on behalf of ERA-EDTA. All rights reserved.
Figures

References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. - PubMed
-
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. - PubMed
-
- Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. - PubMed
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

