Compositional profile of α / β-hydrolase fold proteins in mangrove soil metagenomes: prevalence of epoxide hydrolases and haloalkane dehalogenases in oil-contaminated sites
- PMID: 25171437
- PMCID: PMC4408192
- DOI: 10.1111/1751-7915.12157
Compositional profile of α / β-hydrolase fold proteins in mangrove soil metagenomes: prevalence of epoxide hydrolases and haloalkane dehalogenases in oil-contaminated sites
Abstract
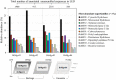
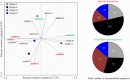
The occurrence of genes encoding biotechnologically relevant α/β-hydrolases in mangrove soil microbial communities was assessed using data obtained by whole-metagenome sequencing of four mangroves areas, denoted BrMgv01 to BrMgv04, in São Paulo, Brazil. The sequences (215 Mb in total) were filtered based on local amino acid alignments against the Lipase Engineering Database. In total, 5923 unassembled sequences were affiliated with 30 different α/β-hydrolase fold superfamilies. The most abundant predicted proteins encompassed cytosolic hydrolases (abH08; ∼ 23%), microsomal hydrolases (abH09; ∼ 12%) and Moraxella lipase-like proteins (abH04 and abH01; < 5%). Detailed analysis of the genes predicted to encode proteins of the abH08 superfamily revealed a high proportion related to epoxide hydrolases and haloalkane dehalogenases in polluted mangroves BrMgv01-02-03. This suggested selection and putative involvement in local degradation/detoxification of the pollutants. Seven sequences that were annotated as genes for putative epoxide hydrolases and five for putative haloalkane dehalogenases were found in a fosmid library generated from BrMgv02 DNA. The latter enzymes were predicted to belong to Actinobacteria, Deinococcus-Thermus, Planctomycetes and Proteobacteria. Our integrated approach thus identified 12 genes (complete and/or partial) that may encode hitherto undescribed enzymes. The low amino acid identity (< 60%) with already-described genes opens perspectives for both production in an expression host and genetic screening of metagenomes.
© 2014 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures

Similar articles
-
Functional metagenomics of oil-impacted mangrove sediments reveals high abundance of hydrolases of biotechnological interest.World J Microbiol Biotechnol. 2017 Jul;33(7):141. doi: 10.1007/s11274-017-2307-5. Epub 2017 Jun 7. World J Microbiol Biotechnol. 2017. PMID: 28593475
-
Gene evolution of epoxide hydrolases and recommended nomenclature.DNA Cell Biol. 1995 Jan;14(1):61-71. doi: 10.1089/dna.1995.14.61. DNA Cell Biol. 1995. PMID: 7832993
-
New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes.Appl Environ Microbiol. 2018 Apr 2;84(8):e02773-17. doi: 10.1128/AEM.02773-17. Print 2018 Apr 15. Appl Environ Microbiol. 2018. PMID: 29427431 Free PMC article.
-
Evolving haloalkane dehalogenases.Curr Opin Chem Biol. 2004 Apr;8(2):150-9. doi: 10.1016/j.cbpa.2004.02.012. Curr Opin Chem Biol. 2004. PMID: 15062775 Review.
-
Epoxide hydrolases: biochemistry and molecular biology.Chem Biol Interact. 2000 Dec 1;129(1-2):41-59. doi: 10.1016/s0009-2797(00)00197-6. Chem Biol Interact. 2000. PMID: 11154734 Review.
Cited by
-
New Thermophilic α/β Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.Front Bioeng Biotechnol. 2018 Oct 16;6:144. doi: 10.3389/fbioe.2018.00144. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30386778 Free PMC article.
-
An Overview on the Enhancement of Enantioselectivity and Stability of Microbial Epoxide Hydrolases.Mol Biotechnol. 2017 Mar;59(2-3):98-116. doi: 10.1007/s12033-017-9996-8. Mol Biotechnol. 2017. PMID: 28271340 Review.
-
Estimating the success of enzyme bioprospecting through metagenomics: current status and future trends.Microb Biotechnol. 2016 Jan;9(1):22-34. doi: 10.1111/1751-7915.12309. Epub 2015 Aug 14. Microb Biotechnol. 2016. PMID: 26275154 Free PMC article. Review.
-
Functional metagenomics of oil-impacted mangrove sediments reveals high abundance of hydrolases of biotechnological interest.World J Microbiol Biotechnol. 2017 Jul;33(7):141. doi: 10.1007/s11274-017-2307-5. Epub 2017 Jun 7. World J Microbiol Biotechnol. 2017. PMID: 28593475
-
The microbial landscape in bioturbated mangrove sediment: A resource for promoting nature-based solutions for mangroves.Microb Biotechnol. 2023 Aug;16(8):1584-1602. doi: 10.1111/1751-7915.14273. Epub 2023 May 20. Microb Biotechnol. 2023. PMID: 37209285 Free PMC article. Review.
References
-
- Arfi Y, Chevret D, Henrissat B, Berrin JG, Levasseur A. Record E. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat Commun. 2013;4:1810. - PubMed
-
- Arulazhagan P. Vasudevan N. Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain Ochrobactrum sp. VA1. Mar Pollut Bull. 2011;62:388–394. - PubMed
-
- Barth S, Fischer M, Schmid RD. Pleiss J. The database of epoxide hydrolases and haloalkane dehalogenases: one structure, many functions. Bioinformatics. 2004;20:2845–2847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

