Epigenetic regulation of female puberty
- PMID: 25171849
- PMCID: PMC6824271
- DOI: 10.1016/j.yfrne.2014.08.003
Epigenetic regulation of female puberty
Abstract
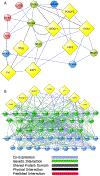
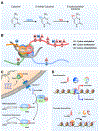
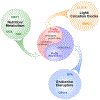
Substantial progress has been made in recent years toward deciphering the molecular and genetic underpinnings of the pubertal process. The availability of powerful new methods to interrogate the human genome has led to the identification of genes that are essential for puberty to occur. Evidence has also emerged suggesting that the initiation of puberty requires the coordinated activity of gene sets organized into functional networks. At a cellular level, it is currently thought that loss of transsynaptic inhibition, accompanied by an increase in excitatory inputs, results in the pubertal activation of GnRH release. This concept notwithstanding, a mechanism of epigenetic repression targeting genes required for the pubertal activation of GnRH neurons was recently identified as a core component of the molecular machinery underlying the central restraint of puberty. In this chapter we will discuss the potential contribution of various mechanisms of epigenetic regulation to the hypothalamic control of female puberty.
Keywords: Chromatin modifications; DNA methylation; Epigenetic regulators; Female puberty; GnRH neurons; Kisspeptin neurons; Long noncoding RNAs; Transcriptional activation; Transcriptional repression; microRNAs.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, Rodrigues T, Kochi C, Longui A, Beckers D, de ZF, Montenegro LR, Mendonca BB, Carroll RS, Hirschhorn JN, Latronico AC, Kaiser UB, 2013. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N. Engl. J. Med 368, 2467–2475. - PMC - PubMed
-
- Ahima RS, Saper CB, Flier JS, Elmquist JK, 2000. Leptin regulation of neuroendocrine systems. Front. Neuroendocrinol 21, 263–307. - PubMed
-
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP, 2002. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol 12, 540–550. - PubMed
-
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub O, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest AR, Carninci P, Rehli M, Sandelin A, Kawaji H, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmid C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple CA, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drablos F, Edge AS, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furuno M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JF, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van NE, Ninomiya N, Nishiyori H, Noma S, Nozaki T, Ogishima S, Ohkura N, Ohmiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JG, Rackham OJ, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Satoh H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, 2014. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

