Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients
- PMID: 25171927
- PMCID: PMC4199956
- DOI: 10.1182/blood-2013-12-546309
Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients
Abstract
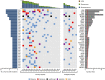
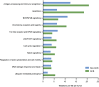
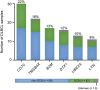
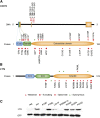
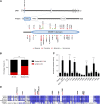
Next-generation sequencing studies on diffuse large B-cell lymphomas (DLBCLs) have revealed novel targets of genetic aberrations but also high intercohort heterogeneity. Previous studies have suggested that the prevalence of disease subgroups and cytogenetic profiles differ between Western and Asian patients. To characterize the coding genome of Chinese DLBCL, we performed whole-exome sequencing of DNA derived from 31 tumors and respective peripheral blood samples. The mutation prevalence of B2M, CD70, DTX1, LYN, TMSB4X, and UBE2A was investigated in an additional 105 tumor samples. We discovered 11 novel targets of recurrent mutations in DLBCL that included functionally relevant genes such as LYN and TMSB4X. Additional genes were found mutated at high frequency (≥10%) in the Chinese cohort including DTX1, which was the most prevalent mutation target in the Notch pathway. We furthermore demonstrated that mutations in DTX1 impair its function as a negative regulator of Notch. Novel and previous unappreciated targets of somatic mutations in DLBCL identified in this study support the existence of additional/alternative tumorigenic pathways in these tumors. The observed differences with previous reports might be explained by the genetic heterogeneity of DLBCL, the germline genetic makeup of Chinese individuals, and/or exposure to distinct etiological agents.
© 2014 by The American Society of Hematology.
Figures

References
-
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008.
-
- Gurbuxani S, Anastasi J, Hyjek E. Diffuse large B-cell lymphoma—more than a diffuse collection of large B cells: an entity in search of a meaningful classification [published correction appears in Arch Pathol Lab Med. 2009;133(8):1186]. Arch Pathol Lab Med. 2009;133(7):1121–1134. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

